Biosynthesis
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
The prerequisite elements for biosynthesis include: precursor compounds, chemical energy (e.g. ATP), and catalytic enzymes which may need coenzymes (e.g. NADH, NADPH). These elements create monomers, the building blocks for macromolecules. Some important biological macromolecules include: proteins, which are composed of amino acid monomers joined via peptide bonds, and DNA molecules, which are composed of nucleotides joined via phosphodiester bonds.
Properties of chemical reactions
Biosynthesis occurs due to a series of chemical reactions. For these reactions to take place, the following elements are necessary:[1]
- Precursor compounds: these compounds are the starting molecules or substrates in a reaction. These may also be viewed as the reactants in a given chemical process.
- Chemical energy: chemical energy can be found in the form of high energy molecules. These molecules are required for energetically unfavorable reactions. Furthermore, the hydrolysis of these compounds drives a reaction forward. High energy molecules, such as ATP, have three phosphates. Often, the terminal phosphate is split off during hydrolysis and transferred to another molecule.
- Catalysts: these may be for example metal ions or coenzymes and they catalyze a reaction by increasing the rate of the reaction and lowering the activation energy.
In the simplest sense, the reactions that occur in biosynthesis have the following format:[2]
Some variations of this basic equation which will be discussed later in more detail are:[3]
- Simple compounds which are converted into other compounds, usually as part of a multiple step reaction pathway. Two examples of this type of reaction occur during the formation of nucleic acids and the charging of tRNA prior to translation. For some of these steps, chemical energy is required:
- Simple compounds that are converted into other compounds with the assistance of cofactors. For example, the synthesis of phospholipids requires acetyl CoA, while the synthesis of another membrane component, sphingolipids, requires NADH and FADH for the formation the sphingosine backbone. The general equation for these examples is:
- Simple compounds that join to create a macromolecule. For example, fatty acids join to form phospholipids. In turn, phospholipids and cholesterol interact noncovalently in order to form the lipid bilayer. This reaction may be depicted as follows:
Lipid

Many intricate macromolecules are synthesized in a pattern of simple, repeated structures.[4] For example, the simplest structures of lipids are fatty acids. Fatty acids are hydrocarbon derivatives; they contain a carboxyl group "head" and a hydrocarbon chain "tail".[4] These fatty acids create larger components, which in turn incorporate noncovalent interactions to form the lipid bilayer.[4] Fatty acid chains are found in two major components of membrane lipids: phospholipids and sphingolipids. A third major membrane component, cholesterol, does not contain these fatty acid units.[5]
Phospholipids
The foundation of all biomembranes consists of a bilayer structure of phospholipids.[6] The phospholipid molecule is amphipathic; it contains a hydrophilic polar head and a hydrophobic nonpolar tail.[4] The phospholipid heads interact with each other and aqueous media, while the hydrocarbon tails orient themselves in the center, away from water.[7] These latter interactions drive the bilayer structure that acts as a barrier for ions and molecules.[8]
There are various types of phospholipids; consequently, their synthesis pathways differ. However, the first step in phospholipid synthesis involves the formation of phosphatidate or diacylglycerol 3-phosphate at the endoplasmic reticulum and outer mitochondrial membrane.[7] The synthesis pathway is found below:

The pathway starts with glycerol 3-phosphate, which gets converted to lysophosphatidate via the addition of a fatty acid chain provided by acyl coenzyme A.[9] Then, lysophosphatidate is converted to phosphatidate via the addition of another fatty acid chain contributed by a second acyl CoA; all of these steps are catalyzed by the glycerol phosphate acyltransferase enzyme.[9] Phospholipid synthesis continues in the endoplasmic reticulum, and the biosynthesis pathway diverges depending on the components of the particular phospholipid.[9]
Sphingolipids
Like phospholipids, these fatty acid derivatives have a polar head and nonpolar tails.[5] Unlike phospholipids, sphingolipids have a sphingosine backbone.[10] Sphingolipids exist in eukaryotic cells and are particularly abundant in the central nervous system.[7] For example, sphingomyelin is part of the myelin sheath of nerve fibers.[11]
Sphingolipids are formed from ceramides that consist of a fatty acid chain attached to the amino group of a sphingosine backbone. These ceramides are synthesized from the acylation of sphingosine.[11] The biosynthetic pathway for sphingosine is found below:

As the image denotes, during sphingosine synthesis, palmitoyl CoA and serine undergo a condensation reaction which results in the formation of 3-dehydrosphinganine.[7] This product is then reduced to form dihydrospingosine, which is converted to sphingosine via the oxidation reaction by FAD.[7]
Cholesterol
This lipid belongs to a class of molecules called sterols.[5] Sterols have four fused rings and a hydroxyl group.[5] Cholesterol is a particularly important molecule. Not only does it serve as a component of lipid membranes, it is also a precursor to several steroid hormones, including cortisol, testosterone, and estrogen.[12]
Cholesterol is synthesized from acetyl CoA.[12] The pathway is shown below:

More generally, this synthesis occurs in three stages, with the first stage taking place in the cytoplasm and the second and third stages occurring in the endoplasmic reticulum.[9] The stages are as follows:[12]
- 1. The synthesis of isopentenyl pyrophosphate, the "building block" of cholesterol
- 2. The formation of squalene via the condensation of six molecules of isopentenyl phosphate
- 3. The conversion of squalene into cholesterol via several enzymatic reactions
Nucleotides
The biosynthesis of nucleotides involves enzyme-catalyzed reactions that convert substrates into more complex products.[1] Nucleotides are the building blocks of DNA and RNA. Nucleotides are composed of a five-membered ring formed from ribose sugar in RNA, and deoxyribose sugar in DNA; these sugars are linked to a purine or pyrimidine base with a glycosidic bond and a phosphate group at the 5' location of the sugar.[13]
Purine nucleotides
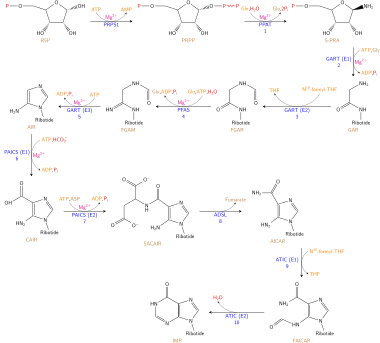
The DNA nucleotides adenosine and guanosine consist of a purine base attached to a ribose sugar with a glycosidic bond. In the case of RNA nucleotides deoxyadenosine and deoxyguanosine, the purine bases are attached to a deoxyribose sugar with a glycosidic bond. The purine bases on DNA and RNA nucleotides are synthesized in a twelve-step reaction mechanism present in most single-celled organisms. Higher eukaryotes employ a similar reaction mechanism in ten reaction steps. Purine bases are synthesized by converting phosphoribosyl pyrophosphate (PRPP) to inosine monophosphate (IMP), which is the first key intermediate in purine base biosynthesis.[14] Further enzymatic modification of IMP produces the adenosine and guanosine bases of nucleotides.
- The first step in purine biosynthesis is a condensation reaction, performed by glutamine-PRPP amidotransferase. This enzyme transfers the amino group from glutamine to PRPP, forming 5-phosphoribosylamine. The following step requires the activation of glycine by the addition of a phosphate group from ATP.
- GAR synthetase[15] performs the condensation of activated glycine onto PRPP, forming glycineamide ribonucleotide (GAR).
- GAR transformylase adds a formyl group onto the amino group of GAR, forming formylglycinamide ribonucleotide (FGAR).
- FGAR amidotransferase[16] catalyzes the addition of a nitrogen group to FGAR, forming formylglycinamidine ribonucleotide (FGAM).
- FGAM cyclase catalyzes ring closure, which involves removal of a water molecule, forming the 5-membered imidazole ring 5-aminoimidazole ribonucleotide (AIR).
- N5-CAIR synthetase transfers a carboxyl group, forming the intermediate N5-carboxyaminoimidazole ribonucleotide (N5-CAIR).[17]
- N5-CAIR mutase rearranges the carboxyl functional group and transfers it onto the imidazole ring, forming carboxyamino- imidazole ribonucleotide (CAIR). The two step mechanism of CAIR formation from AIR is mostly found in single celled organisms. Higher eukaryotes contain the enzyme AIR carboxylase,[18] which transfers a carboxyl group directly to AIR imidazole ring, forming CAIR.
- SAICAR synthetase forms a peptide bond between aspartate and the added carboxyl group of the imidazole ring, forming N-succinyl-5-aminoimidazole-4-carboxamide ribonucleotide (SAICAR).
- SAICAR lyase removes the carbon skeleton of the added aspartate, leaving the amino group and forming 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR).
- AICAR transformylase transfers a carbonyl group to AICAR, forming N-formylaminoimidazole- 4-carboxamide ribonucleotide (FAICAR).
- The final step involves the enzyme IMP synthase, which performs the purine ring closure and forms the inosine monophosphate intermediate.[5]
Pyrimidine nucleotides

Other DNA and RNA nucleotide bases that are linked to the ribose sugar via a glycosidic bond are thymine, cytosine and uracil (which is only found in RNA). Uridine monophosphate biosynthesis involves an enzyme that is located in the mitochondrial inner membrane and multifunctional enzymes that are located in the cytosol.[19]
- The first step involves the enzyme carbamoyl phosphate synthase combining glutamine with CO2 in an ATP dependent reaction to form carbamoyl phosphate.
- Aspartate carbamoyltransferase condenses carbamoyl phosphate with aspartate to form uridosuccinate.
- Dihydroorotase performs ring closure, a reaction that loses water, to form dihydroorotate.
- Dihydroorotate dehydrogenase, located within the mitochondrial inner membrane,[19] oxidizes dihydroorotate to orotate.
- Orotate phosphoribosyl hydrolase (OMP pyrophosphorylase) condenses orotate with PRPP to form orotidine-5'-phosphate.
- OMP decarboxylase catalyzes the conversion of orotidine-5'-phosphate to UMP.[20]
After the uridine nucleotide base is synthesized, the other bases, cytosine and thymine are synthesized. Cytosine biosynthesis is a two-step reaction which involves the conversion of UMP to UTP. Phosphate addition to UMP is catalyzed by a kinase enzyme. The enzyme CTP synthase catalyzes the next reaction step: the conversion of UTP to CTP by transferring an amino group from glutamine to uridine; this forms the cytosine base of CTP.[21] The mechanism, which depicts the reaction UTP + ATP + glutamine ⇔ CTP + ADP + glutamate, is below:
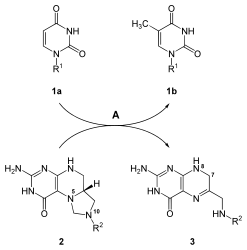

Cytosine is a nucleotide that is present in both DNA and RNA. However, uracil is only found in RNA. Therefore, after UTP is synthesized, it is must be converted into a deoxy form to be incorporated into DNA. This conversion involves the enzyme ribonucleoside triphosphate reductase. This reaction that removes the 2'-OH of the ribose sugar to generate deoxyribose is not affected by the bases attached to the sugar. This non-specificity allows ribonucleoside triphosphate reductase to convert all nucleotide triphosphates to deoxyribonucleotide by a similar mechanism.[21]
In contrast to uracil, thymine bases are found mostly in DNA, not RNA. Cells do not normally contain thymine bases that are linked to ribose sugars in RNA, thus indicating that cells only synthesize deoxyribose-linked thymine. The enzyme thymidylate synthetase is responsible for synthesizing thymine residues from dUMP to dTMP. This reaction transfers a methyl group onto the uracil base of dUMP to generate dTMP.[21] The thymidylate synthase reaction, dUMP + 5,10-methylenetetrahydrofolate ⇔ dTMP + dihydrofolate, is shown to the right.
DNA
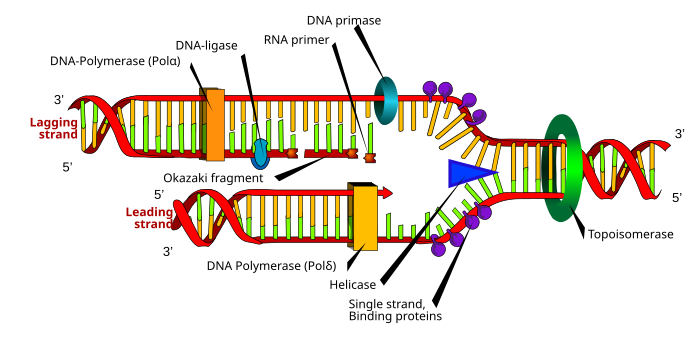
Although there are differences between eukaryotic and prokaryotic DNA synthesis, the following section denotes key characteristics of DNA replication shared by both organisms.
DNA is composed of nucleotides that are joined by phosphodiester bonds.[4] DNA synthesis, which takes place in the nucleus, is a semiconservative process, which means that the resulting DNA molecule contains an original strand from the parent structure and a new strand.[22] DNA synthesis is catalyzed by a family of DNA polymerases that require four deoxynucleoside triphosphates, a template strand, and a primer with a free 3'OH in which to incorporate nucleotides.[23]
In order for DNA replication to occur, a replication fork is created by enzymes called helicases which unwind the DNA helix.[23] Topoisomerases at the replication fork remove supercoils caused by DNA unwinding, and single-stranded DNA binding proteins maintain the two single-stranded DNA templates stabilized prior to replication.[13]
DNA synthesis is initiated by the RNA polymerase primase, which makes an RNA primer with a free 3'OH.[23] This primer is attached to the single-stranded DNA template, and DNA polymerase elongates the chain by incorporating nucleotides; DNA polymerase also proofreads the newly synthesized DNA strand.[23]
During the polymerization reaction catalyzed by DNA polymerase, a nucleophilic attack occurs by the 3'OH of the growing chain on the innermost phosphorus atom of a deoxynucleoside triphosphate; this yields the formation of a phosphodiester bridge that attaches a new nucleotide and releases pyrophosphate.[9]
Two types of strands are created simultaneously during replication: the leading strand, which is synthesized continuously and grows towards the replication fork, and the lagging strand, which is made discontinuously in Okazaki fragments and grows away from the replication fork.[22] Okazaki fragments are covalently joined by DNA ligase to form a continuous strand.[22] Then, to complete DNA replication, RNA primers are removed, and the resulting gaps are replaced with DNA and joined via DNA ligase.[22]
Amino acids
A protein is a polymer that is composed from amino acids that are linked by peptide bonds. There are more than 300 amino acids found in nature of which only twenty, known as the standard amino acids, are the building blocks for protein.[24] Only green plants and most microbes are able to synthesize all of the 20 standard amino acids that are needed by all living species. Mammals can only synthesize ten of the twenty standard amino acids. The other amino acids, valine, methionine, leucine, isoleucine, phenylalanine, lysine, threonine and tryptophan for adults and histidine, and arginine for babies are obtained through diet.[25]
Amino acid basic structure
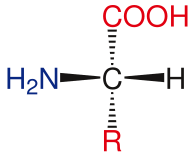
The general structure of the standard amino acids includes a primary amino group, a carboxyl group and the functional group attached to the α-carbon. The different amino acids are identified by the functional group. As a result of the three different groups attached to the α-carbon, amino acids are asymmetrical molecules. For all standard amino acids, except glycine, the α-carbon is a chiral center. In the case of glycine, the α-carbon has two hydrogen atoms, thus adding symmetry to this molecule. With the exception of proline, all of the amino acids found in life have the L-isoform conformation. Proline has a functional group on the α-carbon that forms a ring with the amino group.[24]

Nitrogen source
One major step in amino acid biosynthesis involves incorporating a nitrogen group onto the α-carbon. In cells, there are two major pathways of incorporating nitrogen groups. One pathway involves the enzyme glutamine oxoglutarate aminotransferase (GOGAT) which removes the amide amino group of glutamine and transfers it onto 2-oxoglutarate, producing two glutamate molecules. In this catalysis reaction, glutamine serves as the nitrogen source. An image illustrating this reaction is found to the right.
The other pathway for incorporating nitrogen onto the α-carbon of amino acids involves the enzyme glutamate dehydrogenase (GDH). GDH is able to transfer ammonia onto 2-oxoglutarate and form glutamate. Furthermore, the enzyme glutamine synthetase (GS) is able to transfer ammonia onto glutamate and synthesize glutamine, replenishing glutamine.[26]
The glutamate family of amino acids
The glutamate family of amino acids includes the amino acids that derive from the amino acid glutamate. This family includes: glutamate, glutamine, proline, and arginine. This family also includes the amino acid lysine, which is derived from α-ketoglutarate.[27]
The biosynthesis of glutamate and glutamine is a key step in the nitrogen assimilation discussed above. The enzymes GOGAT and GDH catalyze the nitrogen assimilation reactions.
In bacteria, the enzyme glutamate 5-kinase initiates the biosynthesis of proline by transferring a phosphate group from ATP onto glutamate. The next reaction is catalyzed by the enzyme pyrroline-5-carboxylate synthase (P5CS), which catalyzes the reduction of the ϒ-carboxyl group of L-glutamate 5-phosphate. This results in the formation of glutamate semialdehyde, which spontaneously cyclizes to pyrroline-5-carboxylate. Pyrroline-5-carboxylate is further reduced by the enzyme pyrroline-5-carboxylate reductase (P5CR) to yield a proline amino acid.[28]
In the first step of arginine biosynthesis in bacteria, glutamate is acetylated by transferring the acetyl group from acetyl-CoA at the N-α position; this prevents spontaneous cyclization. The enzyme N-acetylglutamate synthase (glutamate N-acetyltransferase) is responsible for catalyzing the acetylation step. Subsequent steps are catalyzed by the enzymes N-acetylglutamate kinase, N-acetyl-gamma-glutamyl-phosphate reductase, and acetylornithine/succinyldiamino pimelate aminotransferase and yield the N-acetyl-L-ornithine. The acetyl group of acetylornithine is removed by the enzyme acetylornithinase (AO) or ornithine acetyltransferase (OAT), and this yields ornithine. Then, the enzymes citrulline and argininosuccinate convert ornithine to arginine.[29]
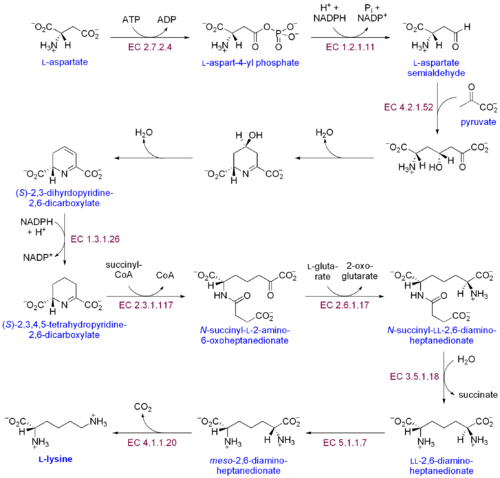
There are two distinct lysine biosynthetic pathways: the diaminopimelic acid pathway and the α-aminoadipate pathway. The most common of the two synthetic pathways is the diaminopimelic acid pathway; it consists of several enzymatic reactions that add carbon groups to aspartate to yield lysine:[30]
- Aspartate kinase initiates the diaminopimelic acid pathway by phosphorylating aspartate and producing aspartyl phosphate.
- Aspartate semialdehyde dehydrogenase catalyzes the NADPH-dependent reduction of aspartyl phosphate to yield aspartate semialdehyde.
- 4-hydroxy-tetrahydrodipicolinate synthase adds a pyruvate group to the β-aspartyl-4-semialdehyde, and a water molecule is removed. This causes cyclization and gives rise to (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate.
- 4-hydroxy-tetrahydrodipicolinate reductase catalyzes the reduction of (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate by NADPH to yield Δ'-piperideine-2,6-dicarboxylate (2,3,4,5-tetrahydrodipicolinate) and H2O.
- Tetrahydrodipicolinate acyltransferase catalyzes the acetylation reaction that results in ring opening and yields N-acetyl α-amino-ε-ketopimelate.
- N-succinyl-α-amino-ε-ketopimelate-glutamate aminotransaminase catalyzes the transamination reaction that removes the keto group of N-acetyl α-amino-ε-ketopimelate and replaces it with an amino group to yield N-succinyl-L-diaminopimelate.[31]
- N-acyldiaminopimelate deacylase catalyzes the deacylation of N-succinyl-L-diaminopimelate to yield L,L-diaminopimelate.[32]
- DAP epimerase catalyzes the conversion of L,L-diaminopimelate to the meso form of L,L-diaminopimelate.[33]
- DAP decarboxylase catalyzes the removal of the carboxyl group, yielding L-lysine.
The serine family of amino acids
The serine family of amino acid includes: serine, cysteine, and glycine. Most microorganisms and plants obtain the sulfur for synthesizing methionine from the amino acid cysteine. Furthermore, the conversion of serine to glycine provides the carbons needed for the biosynthesis of the methionine and histidine.[27]
During serine biosynthesis,[34] the enzyme phosphoglycerate dehydrogenase catalyzes the initial reaction that oxidizes 3-phospho-D-glycerate to yield 3-phosphonooxypyruvate.[35] The following reaction is catalyzed by the enzyme phosphoserine aminotransferase, which transfers an amino group from glutamate onto 3-phosphonooxypyruvate to yield L-phosphoserine.[36] The final step is catalyzed by the enzyme phosphoserine phosphatase, which dephosphorylates L-phosphoserine to yield L-serine.[37]
There are two known pathways for the biosynthesis of glycine. Organisms that use ethanol and acetate as the major carbon source utilize the glyconeogenic pathway to synthesize glycine. The other pathway of glycine biosynthesis is known as the glycolytic pathway. This pathway converts serine synthesized from the intermediates of glycolysis to glycine. In the glycolytic pathway, the enzyme serine hydroxymethyltransferase catalyzes the cleavage of serine to yield glycine and transfers the cleaved carbon group of serine onto tetrahydrofolate, forming 5,10-methylene-tetrahydrofolate.[38]
Cysteine biosynthesis is a two-step reaction that involves the incorporation of inorganic sulfur. In microorganisms and plants, the enzyme serine acetyltransferase catalyzes the transfer of acetyl group from acetyl-CoA onto L-serine to yield O-acetyl-L-serine.[39] The following reaction step, catalyzed by the enzyme O-acetyl serine (thiol) lyase, replaces the acetyl group of O-acetyl-L-serine with sulfide to yield cysteine.[40]
The aspartate family of amino acids
The aspartate family of amino acids includes: threonine, lysine, methionine, isoleucine, and aspartate. Lysine and isoleucine are considered part of the aspartate family even though part of their carbon skeleton is derived from pyruvate. In the case of methionine, the methyl carbon is derived from serine and the sulfur group, but in most organisms, it is derived from cysteine.[27]
The biosynthesis of aspartate is a one step reaction that is catalyzed by a single enzyme. The enzyme aspartate aminotransferase catalyzes the transfer of an amino group from aspartate onto α-ketoglutarate to yield glutamate and oxaloacetate.[41] Asparagine is synthesized by an ATP-dependent addition of an amino group onto aspartate; asparagine synthetase catalyzes the addition of nitrogen from glutamine or soluble ammonia to aspartate to yield asparagine.[42]

The diaminopimelic acid biosynthetic pathway of lysine belongs to the aspartate family of amino acids. This pathway involves nine enzyme-catalyzed reactions that convert aspartate to lysine.[43]
- Aspartate kinase catalyzes the initial step in the diaminopimelic acid pathway by transferring a phosphoryl from ATP onto the carboxylate group of aspartate, which yields aspartyl-β-phosphate.[44]
- Aspartate-semialdehyde dehydrogenase catalyzes the reduction reaction by dephosphorylation of aspartyl-β-phosphate to yield aspartate-β-semialdehyde.[45]
- Dihydrodipicolinate synthase catalyzes the condensation reaction of aspartate-β-semialdehyde with pyruvate to yield dihydrodipicolinic acid.[46]
- 4-hydroxy-tetrahydrodipicolinate reductase catalyzes the reduction of dihydrodipicolinic acid to yield tetrahydrodipicolinic acid.[47]
- Tetrahydrodipicolinate N-succinyltransferase catalyzes the transfer of a succinyl group from succinyl-CoA on to tetrahydrodipicolinic acid to yield N-succinyl-L-2,6-diaminoheptanedioate.[48]
- N-succinyldiaminopimelate aminotransferase catalyzes the transfer of an amino group from glutamate onto N-succinyl-L-2,6-diaminoheptanedioate to yield N-succinyl-L,L-diaminopimelic acid.[49]
- Succinyl-diaminopimelate desuccinylase catalyzes the removal of acyl group from N-succinyl-L,L-diaminopimelic acid to yield L,L-diaminopimelic acid.[50]
- Diaminopimelate epimerase catalyzes the inversion of the α-carbon of L,L-diaminopimelic acid to yield meso-diaminopimelic acid.[51]
- Siaminopimelate decarboxylase catalyzes the final step in lysine biosynthesis that removes the carbon dioxide group from meso-diaminopimelic acid to yield L-lysine.[52]
Proteins
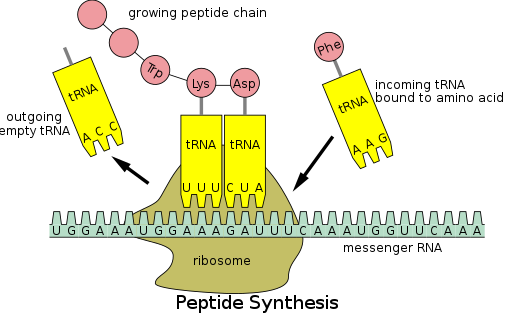
Protein synthesis occurs via a process called translation.[53] During translation, genetic material called mRNA is read by ribosomes to generate a protein polypeptide chain.[53] This process requires transfer RNA (tRNA) which serves as an adaptor by binding amino acids on one end and interacting with mRNA at the other end; the latter pairing between the tRNA and mRNA ensures that the correct amino acid is added to the chain.[53] Protein synthesis occurs in three phases: initiation, elongation, and termination.[13] Prokaryotic (archaeal and bacterial) translation differs from eukaryotic translation; however, this section will mostly focus on the commonalities between the two organisms.
Additional background
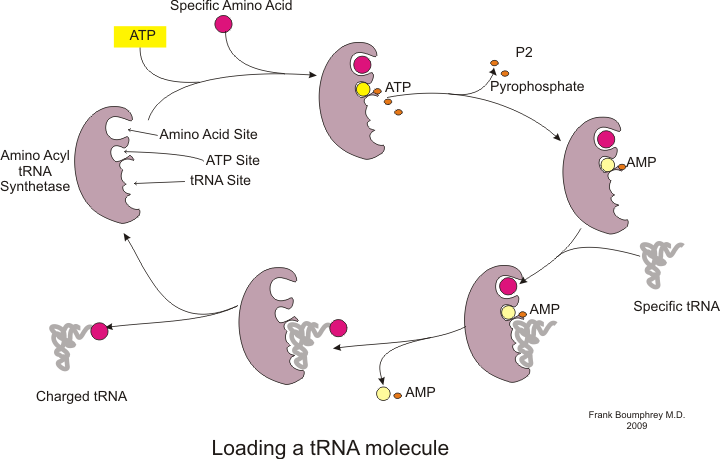
Before translation can begin, the process of binding a specific amino acid to its corresponding tRNA must occur. This reaction, called tRNA charging, is catalyzed by aminoacyl tRNA synthetase.[54] A specific tRNA synthetase is responsible for recognizing and charging a particular amino acid.[54] Furthermore, this enzyme has special discriminator regions to ensure the correct binding between tRNA and its cognate amino acid.[54] The first step for joining an amino acid to its corresponding tRNA is the formation of aminoacyl-AMP:[54]
This is followed by the transfer of the aminoacyl group from aminoacyl-AMP to a tRNA molecule. The resulting molecule is aminoacyl-tRNA:[54]
The combination of these two steps, both of which are catalyzed by aminoacyl tRNA synthetase, produces a charged tRNA that is ready to add amino acids to the growing polypeptide chain.
In addition to binding an amino acid, tRNA has a three nucleotide unit called an anticodon that base pairs with specific nucleotide triplets on the mRNA called codons; codons encode a specific amino acid.[55] This interaction is possible thanks to the ribosome, which serves as the site for protein synthesis. The ribosome possesses three tRNA binding sites: the aminoacyl site (A site), the peptidyl site (P site), and the exit site (E site).[56]
There are numerous codons within an mRNA transcript, and it is very common for an amino acid to be specified by more than one codon; this phenomenon is called degeneracy.[57] In all, there are 64 codons, 61 of each code for one of the 20 amino acids, while the remaining codons specify chain termination.[57]
Translation in steps
As previously mentioned, translation occurs in three phases: initiation, elongation, and termination.

Step 1: Initiation
The completion of the initiation phase is dependent on the following three events:[13]
1. The recruitment of the ribosome to mRNA
2. The binding of a charged initiator tRNA into the P site of the ribosome
3. The proper alignment of the ribosome with mRNA's start codon
Step 2: Elongation
Following initiation, the polypeptide chain is extended via anticodon:codon interactions, with the ribosome adding amino acids to the polypeptide chain one at a time. The following steps must occur to ensure the correct addition of amino acids:[58]
1. The binding of the correct tRNA into the A site of the ribosome
2. The formation of a peptide bond between the tRNA in the A site and the polypeptide chain attached to the tRNA in the P site
3. Translocation or advancement of the tRNA-mRNA complex by three nucleotides
Translocation "kicks off" the tRNA at the E site and shifts the tRNA from the A site into the P site, leaving the A site free for an incoming tRNA to add another amino acid.
Step 3: Termination
The last stage of translation occurs when a stop codon enters the A site.[1] Then, the following steps occur:
1. The recognition of codons by release factors, which causes the hydrolysis of the polypeptide chain from the tRNA located in the P site[1]
2. The release of the polypeptide chain[57]
3. The dissociation and "recycling" of the ribosome for future translation processes[57]
A summary table of the key players in translation is found below:
| Key players in Translation | Translation Stage | Purpose |
|---|---|---|
| tRNA synthetase | before initiation | Responsible for tRNA charging |
| mRNA | initiation, elongation, termination | Template for protein synthesis; contains regions named codons which encode amino acids |
| tRNA | initiation, elongation, termination | Binds ribosomes sites A, P, E; anticodon base pairs with mRNA codon to ensure that the correct amino acid is incorporated into the growing polypeptide chain |
| ribosome | initiation, elongation, termination | Directs protein synthesis and catalyzes the formation of the peptide bond |
Diseases associated with macromolecule deficiency

Errors in biosynthetic pathways can have deleterious consequences including the malformation of macromolecules or the underproduction of functional molecules. Below are examples that illustrate the disruptions that occur due to these inefficiencies.
- Familial hypercholesterolemia: this disorder is characterized by the absence of functional receptors for LDL.[59] Deficiencies in the formation of LDL receptors may cause faulty receptors which disrupt the endocytic pathway, inhibiting the entry of LDL into the liver and other cells.[59] This causes a buildup of LDL in the blood plasma, which results in atherosclerotic plaques that narrow arteries and increase the risk of heart attacks.[59]
- Lesch–Nyhan syndrome: this genetic disease is characterized by self- mutilation, mental deficiency, and gout.[60] It is caused by the absence of hypoxanthine-guanine phosphoribosyltransferase, which is a necessary enzyme for purine nucleotide formation.[60] The lack of enzyme reduces the level of necessary nucleotides and causes the accumulation of biosynthesis intermediates, which results in the aforementioned unusual behavior.[60]
- Severe combined immunodeficiency (SCID): SCID is characterized by a loss of T cells.[61] Shortage of these immune system components increases the susceptibility to infectious agents because the affected individuals cannot develop immunological memory.[61] This immunological disorder results from a deficiency in adenosine deaminase activity, which causes a buildup of dATP. These dATP molecules then inhibit ribonucleotide reductase, which prevents of DNA synthesis.[61]
- Huntington's disease: this neurological disease is caused from errors that occur during DNA synthesis.[62] These errors or mutations lead to the expression of a mutant huntingtin protein, which contains repetitive glutamine residues that are encoded by expanding CAG trinucleotide repeats in the gene.[62] Huntington's disease is characterized by neuronal loss and gliosis. Symptoms of the disease include: movement disorder, cognitive decline, and behavioral disorder.[63]
See also
- Lipids
- Phospholipid bilayer
- Nucleotides
- DNA
- DNA replication
- Proteinogenic amino acid
- Codon table
- Prostaglandin
- Porphyrins
- Chlorophylls and bacteriochlorophylls
- Vitamin B12
References
- ^ a b c d Alberts, Bruce (2008). Molecular Biology of the Cell (5th ed.). New York: Garland Science. ISBN 978-0815341055.
- ^ Zumdahl, Steven S. Zumdahl, Susan A. (2008). Chemistry (8th ed.). CA: Cengage Learning. ISBN 978-0547125329.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Voet, Donald; Voet, Judith G.; Pratt, Charlotte W. (2013). Fundamentals of Biochemistry: Life at the Molecular Level (4th ed.). Hoboken, NJ: Wiley. ISBN 978-0470547847.
- ^ a b c d e Lodish, Harvey; et al. (2007). Molecular cell biology (6th ed.). New York: W.H. Freeman. ISBN 978-0716743668.
- ^ a b c d e Cox, David L. Nelson, Michael M. (2008). Lehninger principles of biochemistry (5th ed.). New York: W.H. Freeman. ISBN 9780716771081.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Hanin, Israel (2013). Phospholipids: Biochemical, Pharmaceutical, and Analytical Considerations. Springer. ISBN 978-1475713664.
- ^ a b c d e Vance, Dennis E.; Vance, Jean E. (2008). Biochemistry of lipids, lipoproteins and membranes (5th ed.). Amsterdam: Elsevier. ISBN 978-0444532190.
- ^ Katsaras, J.; et al. (2001). Lipid bilayers : structure and interactions ; with 6 tables. Berlin [u.a.]: Springer. ISBN 978-3540675556.
- ^ a b c d e Stryer, Jeremy M. Berg; John L. Tymoczko; Lubert (2007). Biochemistry (6. ed., 3. print. ed.). New York: Freeman. ISBN 978-0716787242.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Gault, CR; LM Obeid; YA Hannun (2010). "An Overview of Sphingolipid Metabolism: From Synthesis to Breakdown". Sphingolipids as Signaling and Regulatory Molecules. Advances in Experimental Medicine and Biology. Vol. 688. pp. 1–23. doi:10.1007/978-1-4419-6741-1_1. ISBN 978-1-4419-6740-4. PMC 3069696. PMID 20919643.
- ^ a b Siegel, George J. (1999). Basic neurochemistry : molecular, cellular and medical aspects (6. ed.). Philadelphia, Pa. [u.a.]: Lippincott Williams & Wilkins. ISBN 978-0397518203.
- ^ a b c Harris, J. Robin (2010). Cholesterol binding and cholesterol transport proteins : structure and function in health and disease. Dordrecht: Springer. ISBN 978-9048186211.
- ^ a b c d Watson, James D.; et al. (2007). Molecular biology of the gene (6th ed.). San Francisco, Calif.: Benjamin Cummings. ISBN 978-0805395921.
- ^ Kappock, TJ; Ealick, SE; Stubbe, J (October 2000). "Modular evolution of the purine biosynthetic pathway". Current Opinion in Chemical Biology. 4 (5): 567–72. doi:10.1016/s1367-5931(00)00133-2. PMID 11006546.
- ^ Sampei, G; Baba, S; Kanagawa, M; Yanai, H; Ishii, T; Kawai, H; Fukai, Y; Ebihara, A; Nakagawa, N; Kawai, G (October 2010). "Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria". Journal of Biochemistry. 148 (4): 429–38. doi:10.1093/jb/mvq088. PMID 20716513.
- ^ Hoskins, AA; Anand, R; Ealick, SE; Stubbe, J (Aug 17, 2004). "The formylglycinamide ribonucleotide amidotransferase complex from Bacillus subtilis: metabolite-mediated complex formation". Biochemistry. 43 (32): 10314–27. doi:10.1021/bi049127h. PMID 15301530.
- ^ Mueller, EJ; Meyer, E; Rudolph, J; Davisson, VJ; Stubbe, J (Mar 1, 1994). "N5-carboxyaminoimidazole ribonucleotide: evidence for a new intermediate and two new enzymatic activities in the de novo purine biosynthetic pathway of Escherichia coli". Biochemistry. 33 (8): 2269–78. doi:10.1021/bi00174a038. PMID 8117684.
- ^ Firestine, SM; Poon, SW; Mueller, EJ; Stubbe, J; Davisson, VJ (Oct 4, 1994). "Reactions catalyzed by 5-aminoimidazole ribonucleotide carboxylases from Escherichia coli and Gallus gallus: a case for divergent catalytic mechanisms". Biochemistry. 33 (39): 11927–34. doi:10.1021/bi00205a031. PMID 7918411.
- ^ a b Srere, PA (1987). "Complexes of sequential metabolic enzymes". Annual Review of Biochemistry. 56 (1): 89–124. doi:10.1146/annurev.bi.56.070187.000513. PMID 2441660.
- ^ Broach, edited by Jeffrey N. Strathern, Elizabeth W. Jones, James R. (1981). The Molecular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory. ISBN 978-0879691394.
{{cite book}}:|first=has generic name (help)CS1 maint: multiple names: authors list (link) - ^ a b c O'Donovan, GA; Neuhard, J (September 1970). "Pyrimidine metabolism in microorganisms". Bacteriological Reviews. 34 (3): 278–343. doi:10.1128/MMBR.34.3.278-343.1970. PMC 378357. PMID 4919542.
- ^ a b c d Geer, Gerald Karp ; responsible for the revision of chapter 15 Peter van der (2004). Cell and molecular biology : concepts and experiments (4th ed., Wiley International ed.). New York: J. Wiley & Sons. ISBN 978-0471656654.
{{cite book}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ a b c d Griffiths, Anthony J. F. (1999). Modern genetic analysis (2. print. ed.). New York: Freeman. ISBN 978-0716731184.
- ^ a b Wu, G (May 2009). "Amino acids: metabolism, functions, and nutrition". Amino Acids. 37 (1): 1–17. doi:10.1007/s00726-009-0269-0. PMID 19301095. S2CID 1870305.
- ^ Mousdale, D. M.; Coggins, J. R. (1991). "Amino Acid Synthesis". Target Sites for Herbicide Action. pp. 29–56. doi:10.1007/978-1-4899-2433-9_2. ISBN 978-1-4899-2435-3.
- ^ Miflin, B. J.; Lea, P. J. (1977). "Amino Acid Metabolism". Annual Review of Plant Physiology. 28: 299–329. doi:10.1146/annurev.pp.28.060177.001503.
- ^ a b c Umbarger, HE (1978). "Amino acid biosynthesis and its regulation". Annual Review of Biochemistry. 47 (1): 532–606. doi:10.1146/annurev.bi.47.070178.002533. PMID 354503.
- ^ Pérez-Arellano, I; Carmona-Alvarez, F; Martínez, AI; Rodríguez-Díaz, J; Cervera, J (March 2010). "Pyrroline-5-carboxylate synthase and proline biosynthesis: from osmotolerance to rare metabolic disease". Protein Science. 19 (3): 372–82. doi:10.1002/pro.340. PMC 2866264. PMID 20091669.
- ^ Xu, Y; Labedan, B; Glansdorff, N (March 2007). "Surprising arginine biosynthesis: a reappraisal of the enzymology and evolution of the pathway in microorganisms". Microbiology and Molecular Biology Reviews. 71 (1): 36–47. doi:10.1128/MMBR.00032-06. PMC 1847373. PMID 17347518.
- ^ "MetaCyc: L-lysine biosynthesis I".
- ^ PETERKOFSKY, B; GILVARG, C (May 1961). "N-Succinyl-L-diaminopimelic-glutamic transaminase". The Journal of Biological Chemistry. 236 (5): 1432–8. doi:10.1016/S0021-9258(18)64192-4. PMID 13734750.
- ^ KINDLER, SH; GILVARG, C (December 1960). "N-Succinyl-L-2,6-diaminopimelic acid deacylase". The Journal of Biological Chemistry. 235: 3532–5. doi:10.1016/S0021-9258(18)64502-8. PMID 13756049.
- ^ Born, TL; Blanchard, JS (October 1999). "Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis". Current Opinion in Chemical Biology. 3 (5): 607–13. doi:10.1016/s1367-5931(99)00016-2. PMID 10508663.
- ^ "Escherichia coli K-12 substr. MG1655". serine biosynthesis. SRI International. Retrieved 12 December 2013.
- ^ Bell, JK; Grant, GA; Banaszak, LJ (Mar 30, 2004). "Multiconformational states in phosphoglycerate dehydrogenase". Biochemistry. 43 (12): 3450–8. doi:10.1021/bi035462e. PMID 15035616.
- ^ Dubnovitsky, AP; Kapetaniou, EG; Papageorgiou, AC (January 2005). "Enzyme adaptation to alkaline pH: atomic resolution (1.08 A) structure of phosphoserine aminotransferase from Bacillus alcalophilus". Protein Science. 14 (1): 97–110. doi:10.1110/ps.041029805. PMC 2253317. PMID 15608117.
- ^ Wang, W; Kim, R; Jancarik, J; Yokota, H; Kim, SH (Jan 10, 2001). "Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution". Structure. 9 (1): 65–71. doi:10.1016/s0969-2126(00)00558-x. PMID 11342136.
- ^ Monschau, N; Stahmann, KP; Sahm, H; McNeil, JB; Bognar, AL (May 1, 1997). "Identification of Saccharomyces cerevisiae GLY1 as a threonine aldolase: a key enzyme in glycine biosynthesis". FEMS Microbiology Letters. 150 (1): 55–60. doi:10.1111/j.1574-6968.1997.tb10349.x. PMID 9163906.
- ^ Pye, VE; Tingey, AP; Robson, RL; Moody, PC (Sep 24, 2004). "The structure and mechanism of serine acetyltransferase from Escherichia coli". The Journal of Biological Chemistry. 279 (39): 40729–36. doi:10.1074/jbc.M403751200. PMID 15231846.
- ^ Huang, B; Vetting, MW; Roderick, SL (May 2005). "The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase". Journal of Bacteriology. 187 (9): 3201–5. doi:10.1128/JB.187.9.3201-3205.2005. PMC 1082839. PMID 15838047.
- ^ McPhalen, CA; Vincent, MG; Picot, D; Jansonius, JN; Lesk, AM; Chothia, C (Sep 5, 1992). "Domain closure in mitochondrial aspartate aminotransferase". Journal of Molecular Biology. 227 (1): 197–213. doi:10.1016/0022-2836(92)90691-C. PMID 1522585.
- ^ Larsen, TM; Boehlein, SK; Schuster, SM; Richards, NG; Thoden, JB; Holden, HM; Rayment, I (Dec 7, 1999). "Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product". Biochemistry. 38 (49): 16146–57. CiteSeerX 10.1.1.453.5998. doi:10.1021/bi9915768. PMID 10587437.
- ^ Velasco, AM; Leguina, JI; Lazcano, A (October 2002). "Molecular evolution of the lysine biosynthetic pathways". Journal of Molecular Evolution. 55 (4): 445–59. Bibcode:2002JMolE..55..445V. doi:10.1007/s00239-002-2340-2. PMID 12355264. S2CID 19460256.
- ^ Kotaka, M; Ren, J; Lockyer, M; Hawkins, AR; Stammers, DK (Oct 20, 2006). "Structures of R- and T-state Escherichia coli aspartokinase III. Mechanisms of the allosteric transition and inhibition by lysine". The Journal of Biological Chemistry. 281 (42): 31544–52. doi:10.1074/jbc.M605886200. PMID 16905770.
- ^ Hadfield, A; Kryger, G; Ouyang, J; Petsko, GA; Ringe, D; Viola, R (Jun 18, 1999). "Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis". Journal of Molecular Biology. 289 (4): 991–1002. doi:10.1006/jmbi.1999.2828. PMID 10369777.
- ^ Mirwaldt, C; Korndörfer, I; Huber, R (Feb 10, 1995). "The crystal structure of dihydrodipicolinate synthase from Escherichia coli at 2.5 A resolution". Journal of Molecular Biology. 246 (1): 227–39. doi:10.1006/jmbi.1994.0078. PMID 7853400.
- ^ Cirilli, M; Zheng, R; Scapin, G; Blanchard, JS (Sep 16, 2003). "The three-dimensional structures of the Mycobacterium tuberculosis dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes. Structural and mutagenic analysis of relaxed nucleotide specificity". Biochemistry. 42 (36): 10644–50. doi:10.1021/bi030044v. PMID 12962488.
- ^ Beaman, TW; Binder, DA; Blanchard, JS; Roderick, SL (Jan 21, 1997). "Three-dimensional structure of tetrahydrodipicolinate N-succinyltransferase". Biochemistry. 36 (3): 489–94. doi:10.1021/bi962522q. PMID 9012664.
- ^ Weyand, S; Kefala, G; Weiss, MS (Mar 30, 2007). "The three-dimensional structure of N-succinyldiaminopimelate aminotransferase from Mycobacterium tuberculosis". Journal of Molecular Biology. 367 (3): 825–38. doi:10.1016/j.jmb.2007.01.023. PMID 17292400.
- ^ Nocek, BP; Gillner, DM; Fan, Y; Holz, RC; Joachimiak, A (Apr 2, 2010). "Structural basis for catalysis by the mono- and dimetalated forms of the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase". Journal of Molecular Biology. 397 (3): 617–26. doi:10.1016/j.jmb.2010.01.062. PMC 2885003. PMID 20138056.
- ^ Pillai, B; Cherney, M; Diaper, CM; Sutherland, A; Blanchard, JS; Vederas, JC; James, MN (Nov 23, 2007). "Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase". Biochemical and Biophysical Research Communications. 363 (3): 547–53. doi:10.1016/j.bbrc.2007.09.012. PMID 17889830.
- ^ Gokulan, K; Rupp, B; Pavelka MS, Jr; Jacobs WR, Jr; Sacchettini, JC (May 16, 2003). "Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis". The Journal of Biological Chemistry. 278 (20): 18588–96. doi:10.1074/jbc.M301549200. PMID 12637582.
- ^ a b c Weaver, Robert F. (2005). Molecular biology (3rd ed.). Boston: McGraw-Hill Higher Education. ISBN 978-0-07-284611-9.
- ^ a b c d e Cooper, Geoffrey M. (2000). The Cell: A Molecular Approach (2nd ed.). Washington (DC): ASM Press. ISBN 978-0878931064.
- ^ Jackson, R.J.; et al. (February 2010). "The mechanism of eukaryotic translation initiation and principles of its regulation". Molecular Cell Biology. 10 (2): 113–127. doi:10.1038/nrm2838. PMC 4461372. PMID 20094052.
- ^ Green, Rachel; Harry F. Noller; et al. (1997). "Ribosomes and Translation". Annu. Rev. Biochem. 66: 679–716. doi:10.1146/annurev.biochem.66.1.679. PMID 9242921.
- ^ a b c d Weissbach, Herbert; Pestka, Sidney (1977). Molecular Mechanisms of Protein Biosynthesis. New York: Academic Press. ISBN 978-0127442501.
- ^ Frank, J; Haixiao Gao; et al. (September 2007). "The process of mRNA–tRNA translocation". PNAS. 104 (50): 19671–19678. doi:10.1073/pnas.0708517104. PMC 2148355. PMID 18003906.
- ^ a b c Bandeali, Salman J.; Daye, Jad; Virani, Salim S. (30 November 2013). "Novel Therapies for Treating Familial Hypercholesterolemia". Current Atherosclerosis Reports. 16 (1): 382. doi:10.1007/s11883-013-0382-0. PMID 24293346. S2CID 8903481.
- ^ a b c Kang, Tae Hyuk; Park, Yongjin; Bader, Joel S.; Friedmann, Theodore; Cooney, Austin John (9 October 2013). "The Housekeeping Gene Hypoxanthine Guanine Phosphoribosyltransferase (HPRT) Regulates Multiple Developmental and Metabolic Pathways of Murine Embryonic Stem Cell Neuronal Differentiation". PLOS ONE. 8 (10): e74967. Bibcode:2013PLoSO...874967K. doi:10.1371/journal.pone.0074967. PMC 3794013. PMID 24130677.
- ^ a b c Walport, Ken Murphy, Paul Travers, Mark (2011). Janeway's Immunobiology (8. ed.). Oxford: Taylor & Francis. ISBN 978-0815342434.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b Hughes, edited by Donald C. Lo, Robert E. (2010). Neurobiology of Huntington's disease : applications to drug discovery (2nd ed.). Boca Raton: CRC Press/Taylor & Francis Group. ISBN 978-0849390005.
{{cite book}}:|first=has generic name (help)CS1 maint: multiple names: authors list (link) - ^ Biglan, Kevin M.; Ross, Christopher A.; Langbehn, Douglas R.; Aylward, Elizabeth H.; Stout, Julie C.; Queller, Sarah; Carlozzi, Noelle E.; Duff, Kevin; Beglinger, Leigh J.; Paulsen, Jane S. (26 June 2009). "Motor abnormalities in premanifest persons with Huntington's disease: The PREDICT-HD study". Movement Disorders. 24 (12): 1763–1772. doi:10.1002/mds.22601. PMC 3048804. PMID 19562761.
- CS1 maint: multiple names: authors list
- CS1 errors: generic name
- CS1 maint: numeric names: authors list
- Articles with short description
- Short description is different from Wikidata
- Articles with BNF identifiers
- Articles with BNFdata identifiers
- Articles with GND identifiers
- Articles with J9U identifiers
- Articles with LCCN identifiers
- Articles with LNB identifiers
- Articles with NDL identifiers
- Articles with NKC identifiers
- Articles with EMU identifiers
- Biosynthesis
- Metabolism
![{\displaystyle {\ce {Reactant ->[][enzyme] Product}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/d97fbef58f64fb0b7d05d90fa0a73b05f5919fc8)

![{\displaystyle {\ce {{Precursor~molecule}+Cofactor->[][enzyme]macromolecule}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/b192f25eec9ceebed179bfeb43ae27795f3a91be)


