WRKY protein domain
| WRKY | |||||||||
|---|---|---|---|---|---|---|---|---|---|
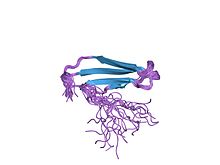 solution structure of the c-terminal wrky domain of atwrky4 | |||||||||
| Identifiers | |||||||||
| Symbol | WRKY | ||||||||
| Pfam | PF03106 | ||||||||
| Pfam clan | CL0274 | ||||||||
| InterPro | IPR003657 | ||||||||
| |||||||||
The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors.[1] The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years.[2] The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) (and variants of this sequence) cis-regulatory element.
Structure
WRKY transcription factors contain either one or two WRKY protein domains. The WRKY protein domain is 60 to 70 amino acids long type of DNA binding domain. The domain is characterized by a highly conserved core WRKYGQK motif and a zinc finger region. The cysteine and histidine zinc finger domain occurs as a CX4-5CX22-23HXH or CX7CX23HXC type, where X can be any amino acid.[3] The zinc finger binds a Zn+2 ion, which is required for protein function.[4] While the WRKYGQK is highly conserved in most WRKY domains, variation in the core sequence has been documented.[2][5] A frequently occurring variant of the core sequence is WRKYGKK, which is present in most plant species.[2][3][5][6][7]
The structure of the WRKY protein domain was first determined in 2005 using nuclear magnetic resonance (NMR) and later by crystallography.[4][8] The WRKY protein domain is a globular shape composed of five anti-parallel β-strands. The core WRKYGQK motif is found on the second β-strand.[8] Eighteen amino acids are highly conserved in the WRKY protein domain, including the core motif, zinc-finger binding cysteines and histidines, and a triad forming a DWK salt bridge.[8] The triad consist of a conserved tryptophan (W) of the core motif, along with an aspartic acid (D) four amino acids upstream and a lysine (K) 29 amino acids downstream of it, stabilizing the entire domain.[8] Five amino acids on the third β-strand (PRSYY) are also well conserved in the WRKY domain.[8] Importantly, the WRKY genes contain a conserved intron in the WRKY domain, which occurs at the location encoding for the PR of the PRSYY amino acid sequence,[3] thus explaining the conservation of this motif.
WRKY-DNA Interaction
The WRKY domain forms a unique wedge-shaped structure that enters perpendicularly in the major groove of the DNA strand.[9] WRKY protein domains interact with the (T/A)TGAC(T/A) cis-element, also called the W-box.[1][10][11] Recent evidence suggests that the GAC core of the W-box is the primary target of the WRKY domain and flanking sequences help dictate DNA interaction with very specific WRKY proteins.[12] The RKYGQK residues of the core motif and additional arginine and lysine residues of the WRKY domain are responsible for interaction with the phosphate backbone of seven consecutive DNA base pairs, including the GAC core.[9][12] Changing the tryptophan, tyrosine, or either lysine of the WRKYGQK motif to alanine completely abolishes DNA-binding,[8][13] indicating these amino acids are essential for recognizing the W-box element. While not essential, altering the WRKYGQK motif arginine, glycine or glutamine to alanine reduces DNA-binding to the W-box.[8][13] Overall, these complex WRKY protein domain-DNA interactions results in gene activation necessary for numerous aspects of plant development and defense.
External links
- WRKY family at PlantTFDB: Plant Transcription Factor Database
- WRKY Transcription Factor Family at The Arabidopsis Information Resource
- The Rushton Lab
- The Somssich Lab
- The Shen Lab
- Somssich’s list of WRKY-related publications
- Eulgem Lab
References
- ^ a b Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (October 1996). "Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes". The EMBO Journal. 15 (20): 5690–700. doi:10.1002/j.1460-2075.1996.tb00953.x. PMC 452313. PMID 8896462.
- ^ a b c Schluttenhofer C, Yuan L (February 2015). "Regulation of specialized metabolism by WRKY transcription factors". Plant Physiology. 167 (2): 295–306. doi:10.1104/pp.114.251769. PMC 4326757. PMID 25501946.
- ^ a b c Eulgem T, Rushton PJ, Robatzek S, Somssich IE (May 2000). "The WRKY superfamily of plant transcription factors". Trends in Plant Science. 5 (5): 199–206. doi:10.1016/s1360-1385(00)01600-9. PMID 10785665.
- ^ a b Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, et al. (March 2005). "Solution structure of an Arabidopsis WRKY DNA binding domain". The Plant Cell. 17 (3): 944–56. doi:10.1105/tpc.104.026435. PMC 1069710. PMID 15705956.
- ^ a b Zhang Y, Wang L (January 2005). "The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants". BMC Evolutionary Biology. 5: 1. doi:10.1186/1471-2148-5-1. PMC 544883. PMID 15629062.
- ^ Song H, Wang P, Nan Z, Wang X (2014). "The WRKY Transcription Factor Genes in Lotus japonicus". International Journal of Genomics. 2014: 420128. doi:10.1155/2014/420128. PMC 3976811. PMID 24745006.
- ^ Xiong W, Xu X, Zhang L, Wu P, Chen Y, Li M, Jiang H, Wu G (July 2013). "Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.)". Gene. 524 (2): 124–32. doi:10.1016/j.gene.2013.04.047. PMID 23644253.
- ^ a b c d e f g Duan MR, Nan J, Liang YH, Mao P, Lu L, Li L, Wei C, Lai L, Li Y, Su XD (2007). "DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein". Nucleic Acids Research. 35 (4): 1145–54. doi:10.1093/nar/gkm001. PMC 1851648. PMID 17264121.
- ^ a b Yamasaki K, Kigawa T, Watanabe S, Inoue M, Yamasaki T, Seki M, Shinozaki K, Yokoyama S (March 2012). "Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor". The Journal of Biological Chemistry. 287 (10): 7683–91. doi:10.1074/jbc.M111.279844. PMC 3293589. PMID 22219184.
- ^ Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE (September 1999). "Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors". The EMBO Journal. 18 (17): 4689–99. doi:10.1093/emboj/18.17.4689. PMC 1171542. PMID 10469648.
- ^ de Pater S, Greco V, Pham K, Memelink J, Kijne J (December 1996). "Characterization of a zinc-dependent transcriptional activator from Arabidopsis". Nucleic Acids Research. 24 (23): 4624–31. doi:10.1093/nar/24.23.4624. PMC 146317. PMID 8972846.
- ^ a b Brand LH, Fischer NM, Harter K, Kohlbacher O, Wanke D (November 2013). "Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays". Nucleic Acids Research. 41 (21): 9764–78. doi:10.1093/nar/gkt732. PMC 3834811. PMID 23975197.
- ^ a b Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K (November 2001). "Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins". Bioscience, Biotechnology, and Biochemistry. 65 (11): 2428–36. doi:10.1271/bbb.65.2428. PMID 11791715. S2CID 22671192.