Polyacrylamide gel electrophoresis

Polyacrylamide gel electrophoresis (PAGE) is a technique widely used in biochemistry, forensic chemistry, genetics, molecular biology and biotechnology to separate biological macromolecules, usually proteins or nucleic acids, according to their electrophoretic mobility. Electrophoretic mobility is a function of the length, conformation, and charge of the molecule. Polyacrylamide gel electrophoresis is a powerful tool used to analyze RNA samples. When polyacrylamide gel is denatured after electrophoresis, it provides information on the sample composition of the RNA species.[1]
Hydration of acrylonitrile results in formation of acrylamide molecules (C
3H
5NO) by nitrile hydratase.[2] Acrylamide monomer is in a powder state before addition of water. Acrylamide is toxic to the human nervous system, therefore all safety measures must be followed when working with it. Acrylamide is soluble in water and upon addition of free-radical initiators it polymerizes resulting in formation of polyacrylamide.[2]
It is useful to make polyacrylamide gel via acrylamide hydration because pore size can be regulated. Increased concentrations of acrylamide result in decreased pore size after polymerization. Polyacrylamide gel with small pores helps to examine smaller molecules better since the small molecules can enter the pores and travel through the gel while large molecules get trapped at the pore openings.[3][4][5]
Purpose and history
The history of electrophoresis for molecular separation and chemical analysis began with the work of Arne Tiselius in 1931, while new separation processes and chemical analysis techniques based on electrophoresis continue to be developed in the 21st century. Tiselius, with support from the Rockefeller Foundation, developed the "Tiselius apparatus" for moving boundary electrophoresis, which was described in 1937 in the well-known paper "A New Apparatus for Electrophoretic Analysis of Colloidal Mixtures".[6] The method spread slowly until the advent of effective zone electrophoresis methods in the 1940s and 1950s, which used filter paper or gels as supporting media. By the 1960s, increasingly sophisticated gel electrophoresis methods made it possible to separate biological molecules based on minute physical and chemical differences, helping to drive the rise of molecular biology. Gel electrophoresis and related techniques became the basis for a wide range of biochemical methods, such as protein fingerprinting, Southern blot, other blotting procedures, DNA sequencing[7][8]
In terms of the purpose of Polyacrylamide gel electrophoresis, we find these are some of the aims:[9]
- PAGE is used to quantify single proteins from a mixture
- It can come from a patient sample or bacterial culture
- Separates DNA and RNA
- Used with technique called western blotting
Methods
As with all forms of gel electrophoresis, molecules may be run in their native state, preserving the molecules' higher-order structure. This method is called native-PAGE. Alternatively, a chemical denaturant may be added to remove this structure and turn the molecule into an unstructured molecule whose mobility depends only on its length (because the protein-SDS complexes all have a similar mass-to-charge ratio). This procedure is called SDS-PAGE. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) is a method of separating molecules based on the difference of their molecular weight. At the pH at which gel electrophoresis is carried out the SDS molecules are negatively charged and bind to proteins in a set ratio, approximately one molecule of SDS for every 2 amino acids.[10]: 164–79 In this way, the detergent provides all proteins with a uniform charge-to-mass ratio. By binding to the proteins the detergent destroys their secondary, tertiary and/or quaternary structure denaturing them and turning them into negatively charged linear polypeptide chains. When subjected to an electric field in PAGE, the negatively charged polypeptide chains travel toward the anode with different mobility. Their mobility, or the distance traveled by molecules, is inversely proportional to the logarithm of their molecular weight.[11] By comparing the relative ratio of the distance traveled by each protein to the length of the gel (Rf) one can make conclusions about the relative molecular weight of the proteins, where the length of the gel is determined by the distance traveled by a small molecule like a tracking dye.[12]For nucleic acids, urea is the most commonly used denaturant. For proteins, sodium dodecyl sulfate (SDS) is an anionic detergent applied to protein samples to coat proteins in order to impart two negative charges (from every SDS molecule) to every two amino acids of the denatured protein.[10]: 161–3 2-Mercaptoethanol may also be used to disrupt the disulfide bonds found between the protein complexes, which helps further denature the protein. In most proteins, the binding of SDS to the polypeptide chains impart an even distribution of charge per unit mass, thereby resulting in a fractionation by approximate size during electrophoresis. Proteins that have a greater hydrophobic content – for instance, many membrane proteins, and those that interact with surfactants in their native environment – are intrinsically harder to treat accurately using this method, due to the greater variability in the ratio of bound SDS.[13] Procedurally, using both Native and SDS-PAGE together can be used to purify and to separate the various subunits of the protein. Native-PAGE keeps the oligomeric form intact and will show a band on the gel that is representative of the level of activity. SDS-PAGE will denature and separate the oligomeric form into its monomers, showing bands that are representative of their molecular weights. These bands can be used to identify and assess the purity of the protein.[10]: 161–3
Procedure
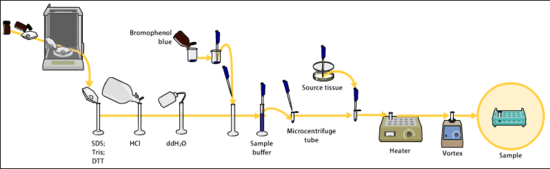
Samples may be any material containing proteins or nucleic acids, these may be biologically derived, for example from prokaryotic or eukaryotic cells, tissues, viruses, environmental samples, or purified proteins. In the case of solid tissues or cells, these are often first broken down mechanically using a blender,or using a homogenizer, and a combination of biochemical and mechanical techniques may be used to separate different cell compartments and organelles prior to electrophoresis.[14][15][16]
The sample to analyze is optionally mixed with a chemical denaturant if so desired, usually SDS for proteins or urea for nucleic acids. SDS is an anionic detergent that denatures secondary and non–disulfide–linked tertiary structures, and additionally applies a negative charge to each protein in proportion to its mass. Urea breaks the hydrogen bonds between the base pairs of the nucleic acid, causing the constituent strands to separate. Heating the samples to at least 60 °C further promotes denaturation.[17][18][19][20]
In addition to SDS, proteins may optionally be briefly heated to near boiling in the presence of a reducing agent, such as 2-mercaptoethanol (beta-mercaptoethanol/BME), which further denatures the proteins by reducing disulfide linkages, thus overcoming some forms of tertiary protein folding, and breaking up quaternary protein structure, this is known as reducing SDS-PAGE [lower-alpha 1][15][21]
Preparing
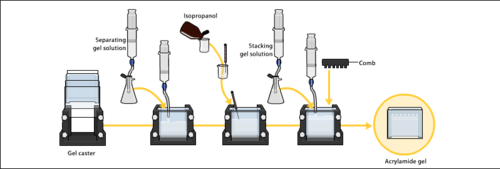
The gels typically consist of acrylamide, bisacrylamide, the optional denaturant (SDS or urea), and a buffer with an adjusted pH. The solution may be degassed under a vacuum to prevent the formation of air bubbles during polymerization. Alternatively, butanol may be added to the resolving gel (for proteins) after it is poured, as butanol removes bubbles and makes the surface smooth. [22] A source of free radicals and a stabilizer, such as ammonium persulfate and TEMED are added to initiate polymerization.[23] The polymerization reaction creates a gel because of the added bisacrylamide, which can form cross-links between two acrylamide molecules. The ratio of bisacrylamide to acrylamide can be varied for special purposes, but is generally about 1 part in 35. The acrylamide concentration of the gel can also be varied, generally in the range from 5% to 25%. Lower percentage gels are better for resolving very high molecular weight molecules, while much higher percentages of acrylamide are needed to resolve smaller proteins. The average pore diameter of polyacrylamide gels is determined by the total concentration of acrylamides (% T with T = Total concentration of acrylamide and bisacrylamide) and the concentration of the cross-linker bisacrylamide (%C with C = bisacrylamide concentration).[24]

Electrophoresis
Various buffer systems are used in PAGE depending on the nature of the sample and the experimental objective. The buffers used at the anode and cathode may be the same or different.[19][25] [26]

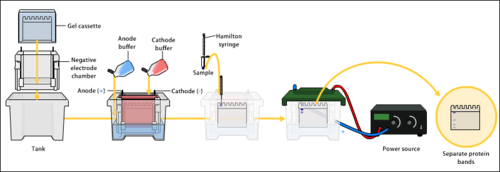
An electric field is applied across the gel, causing the negatively charged proteins or nucleic acids to migrate across the gel away from the negative electrode (which is the cathode being that this is an electrolytic rather than galvanic cell) and towards the positive electrode (the anode). Depending on their size, each biomolecule moves differently through the gel matrix: small molecules more easily fit through the pores in the gel, while larger ones have more difficulty. The gel is run usually for a few hours, though this depends on the voltage applied across the gel; migration occurs more quickly at higher voltages, but these results are typically less accurate than at those at lower voltages. After the set amount of time, the biomolecules have migrated different distances based on their size. Smaller biomolecules travel farther down the gel, while larger ones remain closer to the point of origin. Biomolecules may therefore be separated roughly according to size, which depends mainly on molecular weight under denaturing conditions, but also depends on higher-order conformation under native conditions. The gel mobility is defined as the rate of migration traveled with a voltage gradient of 1V/cm and has units of cm2/sec/V.[10]: 161–3 For analytical purposes, the relative mobility of biomolecules, Rf, the ratio of the distance the molecule traveled on the gel to the total travel distance of a tracking dye is plotted versus the molecular weight of the molecule (or sometimes the log of MW, or rather the Mr, molecular radius). Such typically linear plots represent the standard markers or calibration curves that are widely used for the quantitative estimation of a variety of biomolecular sizes.[10]: 161–3
Certain glycoproteins, however, behave anomalously on SDS gels. Additionally, the analysis of larger proteins ranging from 250,000 to 600,000 Da is also reported to be problematic due to the fact that such polypeptides move improperly in the normally used gel systems.[27]
Further process
Following electrophoresis, the gel may be stained (for proteins, most commonly with Coomassie brilliant blue R-250 ; for nucleic acids, ethidium bromide; or for either, silver stain[28][29][30]), allowing visualization of the separated proteins, or processed further .
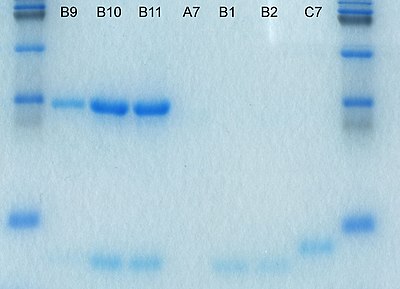
After staining, different species biomolecules appear as distinct bands within the gel. It is common to run molecular weight size markers of known molecular weight in a separate lane in the gel to calibrate the gel and determine the approximate molecular mass of unknown biomolecules by comparing the distance traveled relative to the marker.For proteins, SDS-PAGE is usually the first choice as an assay of purity due to its reliability and ease. The presence of SDS and the denaturing step make proteins separate, approximately based on size, but aberrant migration of some proteins may occur. Different proteins may also stain differently, which interferes with quantification by staining. PAGE may also be used as a preparative technique for the purification of proteins. For example, quantitative preparative native continuous polyacrylamide gel electrophoresis (QPNC-PAGE) is a method for separating native metalloproteins in complex biological matrices.[31][32][33][34]
Components and their roles
Polyacrylamide gel had been known as a potential embedding medium for sectioning tissues as early as 1964, and two independent groups employed PAG in electrophoresis in 1959.[35][36] It possesses several electrophoretically desirable features that make it a versatile medium. It is a synthetic, thermo-stable, transparent, strong, chemically relatively inert gel, and can be prepared with a wide range of average pore sizes.[37]
The pore size of a gel and the reproducibility in gel pore size are determined by two factors, the total amount of acrylamide present (%T) (T = Total concentration of acrylamide and bisacrylamide monomer), and the amount of cross-linker (C = bisacrylamide concentration). Pore size decreases with increasing %T; with cross-linking, 5%C gives the smallest pore size. This gel material can also withstand high voltage gradients, is amenable to various staining and destaining procedures, and can be digested to extract separated fractions or dried for autoradiography and permanent recording.[38][39][4]
Components
Polyacrylamide gels are composed of a stacking gel and separating gel. Stacking gels have a higher porosity relative to the separating gel, and allow for proteins to migrate in a concentrated area. Additionally, stacking gels usually have a pH of 6.8, since the neutral glycine molecules allow for faster protein mobility. Separating gels have a pH of 8.8, where the anionic glycine slows down the mobility of proteins. Separating gels allow for the separation of proteins and have a relatively lower porosity. Here, the proteins are separated based on size (in SDS-PAGE) and size/ charge (Native PAGE).[40]
- Chemical buffer stabilizes the pH value to the desired value within the gel itself and in the electrophoresis buffer. The choice of buffer also affects the electrophoretic mobility of the buffer counterions and thereby the resolution of the gel. The buffer should also be unreactive and not modify or react with most proteins. Different buffers may be used as cathode and anode buffers, respectively, depending on the application. Multiple pH values may be used within a single gel, for example in DISC electrophoresis. Common buffers in PAGE include Tris, Bis-Tris, or imidazole.[41][42][43]
- Counterion balance the intrinsic charge of the buffer ion and also affect the electric field strength during electrophoresis. Highly charged and mobile ions are often avoided in SDS-PAGE cathode buffers, but may be included in the gel itself, where it migrates ahead of the protein. In applications such as DISC SDS-PAGE the pH values within the gel may vary to change the average charge of the counterions during the run to improve resolution. Popular counterions are glycine and tricine, glycine has been used as the source of trailing ion or slow ion because its pKa and mobility of glycinate are such that the effective mobility can be set at a value below that of the slowest known proteins of net negative charge in the pH range.[44][45][46]
- Acrylamide (C
3H
5NO; mW: 71.08) when dissolved in water, slow, spontaneous autopolymerization of acrylamide takes place, joining molecules together by head on tail fashion to form long single-chain polymers. The presence of a free radical-generating system greatly accelerates polymerization. This kind of reaction is known as vinyl addition polymerisation. A solution of these polymer chains becomes viscous but does not form a gel, because the chains simply slide over one another. Gel formation requires linking various chains together. Acrylamide is carcinogenic,[47] a neurotoxin, and a reproductive toxin.[48] - Bisacrylamide (N,N′-Methylenebisacrylamide) (C
7H
10N
2O
2; mW: 154.17) is the most frequently used cross linking agent for polyacrylamide gels. Chemically it can be thought of as two acrylamide molecules coupled head to head at their non-reactive ends. Bisacrylamide can crosslink two polyacrylamide chains to one another, thereby resulting in a gel.[49][50]

- Sodium dodecyl sulfate (SDS) (C
12H
25NaO
4S; mW: 288.38) (only used in denaturing protein gels) is a strong detergent agent used to denature native proteins to individual polypeptides. This denaturation, which is referred to as reconstructive denaturation, is not accomplished by the total linearization of the protein, but instead, through a conformational change to a combination of random coil and α helix secondary structures.[13]
- When a protein mixture is heated to 100 °C in presence of SDS, the detergent wraps around the polypeptide backbone. In this process, the intrinsic charges of polypeptides become negligible when compared to the negative charges contributed by SDS. Thus polypeptides after treatment become rod-like structures possessing a uniform charge density. Without SDS, different proteins with similar molecular weights would migrate differently due to differences in mass-charge ratio, as each protein has an isoelectric point and molecular weight particular to its primary structure. Adding SDS solves this problem, as it binds to and unfolds the protein, giving a near uniform negative charge along the length of the polypeptide.[51][52][53][54]
- Urea (CO(NH
2)
2; mW: 60.06) is a chaotropic agent that increases the entropy of the system by interfering with intramolecular interactions mediated by non-covalent forces such as hydrogen bonds . Macromolecular structure is dependent on the net effect of these forces, therefore it follows that an increase in chaotropic solutes denatures macromolecules.[55][56] - Ammonium persulfate (APS) (N
2H
8S
2O
8; mW: 228.2) is a source of free radicals and is often used as an initiator for gel formation.[57] - TEMED (N, N, N′, N′-tetramethylethylenediamine) (C
6H
16N
2; mW: 116.21) stabilizes free radicals and improves polymerization. The rate of polymerisation and the properties of the resulting gel depend on the concentrations of free radicals. Increasing the amount of free radicals results in a decrease in the average polymer chain length, an increase in gel turbidity and a decrease in gel elasticity. Decreasing the amount shows the reverse effect. The lowest catalytic concentrations that allow polymerisation in a reasonable period of time should be used.[58][59]
Chemicals for processing & visualization
The following chemicals and procedures are used for processing of the gel and the protein samples visualized in it:
- Tracking dye; as proteins and nucleic acids are mostly colorless, their progress through the gel during electrophoresis cannot be easily followed. Anionic dyes of a known electrophoretic mobility are therefore usually included in the PAGE sample buffer. A very common tracking dye is Bromophenol blue. This dye is coloured at alkali and neutral pH and is a small negatively charged molecule that moves towards the anode. Being a highly mobile molecule it moves ahead of most proteins. As it reaches the anodic end of the electrophoresis medium electrophoresis is stopped. It can weakly bind to some proteins and impart a blue colour. Other common tracking dyes are xylene cyanol, which has lower mobility, and Orange G, which has a higher mobility.[60][61][62][63]

- Coomassie brilliant blue R-250 (CBB)(C
45H
44N
3NaO
7S
2; mW: 825.97) is the most popular protein stain. It is an anionic dye, which non-specifically binds to proteins. The structure of CBB is predominantly non-polar, and it is usually used in methanolic solution acidified with acetic acid. Proteins in the gel are fixed by acetic acid and simultaneously stained. The excess dye incorporated into the gel can be removed by destaining with the same solution without the dye. The proteins are detected as blue bands on a clear background. As SDS is also anionic, it may interfere with staining process.[64][28][65] - Ethidium bromide (EtBr) is a popular nucleic acid stain. EtBr allows one to easily visualize DNA or RNA on a gel as EtBr fluoresces an orange color under UV light.[66] Ethidium bromide binds nucleic acid chains through the process of Intercalation.[10] While Ethidium bromide is a popular stain it is important to exercise caution when using EtBr as it is a known carcinogen. Because of this fact, many researchers opt to use stains such as SYBR Green and SYBR Safe which are safer alternatives to EtBr.[67] EtBr is used by simply adding it to the gel mixture. Once the gel has run, the gel may be viewed through the use of a photo-documentation system.[10]

- Silver staining is used when more sensitive method for detection is needed, as classical Coomassie Brilliant Blue staining can usually detect a 50 ng protein band, Silver staining increases the sensitivity typically 10-100 fold more. This is based on the chemistry of photographic development. The proteins are fixed to the gel with a dilute methanol solution, then incubated with an acidic silver nitrate solution. Silver ions are reduced to their metallic form by formaldehyde at alkaline pH. An acidic solution, such as acetic acid stops development.[68] Silver staining was introduced by Kerenyi and Gallyas as a sensitive procedure to detect trace amounts of proteins in gels.[69] The technique has been extended to the study of other biological macromolecules that have been separated in a variety of supports.[70] Many variables can influence the colour intensity and every protein has its own staining characteristics; clean glassware, pure reagents and water of highest purity are the key points to successful staining.[71] Silver staining was developed in the 14th century for colouring the surface of glass. It has been used extensively for this purpose since the 16th century. The colour produced by the early silver stains ranged between light yellow and an orange-red. Camillo Golgi perfected the silver staining for the study of the nervous system. Golgi's method stains a limited number of cells at random in their entirety.[72]
- Autoradiography, also used for protein band detection post gel electrophoresis, uses radioactive isotopes to label proteins, which are then detected by using X-ray film.[73]
- Western blotting is a process by which proteins separated in the acrylamide gel are electrophoretically transferred to a stable, manipulable membrane such as a nitrocellulose, nylon, or PVDF membrane. It is then possible to apply immunochemical techniques to visualise the transferred proteins, as well as accurately identify relative increases or decreases of the protein of interest.[74][75]
See also
- Agarose gel electrophoresis
- Capillary electrophoresis
- DNA electrophoresis
- Eastern blotting
- Electroblotting
- History of electrophoresis
- Isoelectric focusing
- Isotachophoresis
- Native gel electrophoresis
- Northern blotting
- Protein electrophoresis
- QPNC-PAGE
- Southern blotting
- Two dimensional SDS-PAGE
- Zymography
Notes
- ↑ tracking dye may be added to the solution, this typically has a higher electrophoretic mobility than the analytes to allow the experimenter to track the progress of the solution through the gel during the electrophoretic run
References
- ↑ Petrov A, Tsa A, Puglisi JD (2013). "Chapter Sixteen – Analysis of RNA by Analytical Polyacrylamide Gel Electrophoresis". In Lorsch J (ed.). Methods in Enzymology. Vol. 530. Academic Press. pp. 301–313. doi:10.1016/B978-0-12-420037-1.00016-6. ISBN 978-0-12-420037-1. PMID 24034328.
- ↑ 2.0 2.1 The Editors of Encyclopaedia Britannica (2017). "Polyacrylamide". Britannica Online Academic Edition. Encyclopedia Britannica, Inc. Archived from the original on 9 January 2023. Retrieved 2 March 2023.
- ↑ Simpson, Richard J. (April 2010). "Pouring linear gradient gels with a gradient former". Cold Spring Harbor Protocols. 2010 (4): pdb.prot5411. doi:10.1101/pdb.prot5411. ISSN 1559-6095. PMID 20360365. Archived from the original on 1 July 2023. Retrieved 6 May 2023.
- ↑ 4.0 4.1 Green, Michael R.; Sambrook, Joseph (1 December 2020). "Polyacrylamide Gel Electrophoresis". Cold Spring Harbor Protocols. 2020 (12). doi:10.1101/pdb.prot100412. ISSN 1559-6095. Archived from the original on 10 May 2023. Retrieved 25 May 2023.
- ↑ Hendry, G. A.; Grime, J. P. (6 December 2012). Methods in Comparative Plant Ecology: A laboratory manual. Springer Science & Business Media. p. 219. ISBN 978-94-011-1494-3. Archived from the original on 1 July 2023. Retrieved 22 May 2023.
- ↑ Tiselius, Arne (1937). "A new apparatus for electrophoretic analysis of colloidal mixtures". Transactions of the Faraday Society. 33: 524–531. doi:10.1039/TF9373300524.
- ↑ Suárez-Díaz, Edna (September 2022). "The Electrophoretic Revolution in the 1960s: Historical Epistemology Meets the Global History of Science and Technology". Berichte Zur Wissenschaftsgeschichte. 45 (3): 332–343. doi:10.1002/bewi.202200024. ISSN 1522-2365. Archived from the original on 18 February 2023. Retrieved 11 May 2023.
- ↑ Smithies, Oliver (2012). "How it all began: a personal history of gel electrophoresis". Methods in Molecular Biology (Clifton, N.J.). 869: 1–21. doi:10.1007/978-1-61779-821-4_1. ISSN 1940-6029. Archived from the original on 15 June 2022. Retrieved 15 May 2023.
- ↑ Stringer, R. (1 January 2005). "ELECTROPHORESIS | Overview". Encyclopedia of Analytical Science (Second ed.). Elsevier. pp. 356–363. ISBN 978-0-12-369397-6.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 Ninfa AJ, Ballou DP, Benore M (2010). Fundamental Laboratory Approaches for Biochemistry and Biotechnology (2nd ed.). Hoboken, NJ: John Wiley & Sons, Inc. ISBN 978-0-470-08766-4. OCLC 420027217.
- ↑ Kindt T, Goldsby R, Osborne B (2007). Kuby Immunology. New York: W.H. Freeman and Company. p. 553. ISBN 978-1-4292-0211-4.
- ↑ Kumar A, Awasthi A (2009). Bioseparation Engineering. New Delhi: I.K. International Publishing House. p. 137. ISBN 9789380026084.
- ↑ 13.0 13.1 Rath A, Glibowicka M, Nadeau VG, et al. (2009). "Detergent binding explains anomalous SDS-PAGE migration of membrane proteins". Proc. Natl. Acad. Sci. U.S.A. 106 (6): 1760–5. Bibcode:2009PNAS..106.1760R. doi:10.1073/pnas.0813167106. PMC 2644111. PMID 19181854.
- ↑ "Homogenizers for Mixing, Dispersing, and Emulsifying". proscientific.com. Archived from the original on 17 April 2023. Retrieved 21 May 2023.
- ↑ 15.0 15.1 "A guide to Polyacrylamide gel electrophoresis and detection" (PDF). Bio-Rad. Archived (PDF) from the original on 8 May 2022. Retrieved 7 May 2023.
- ↑ Garcia-Vaquero, M.; Rajauria, G.; O'Doherty, J. V.; Sweeney, T. (1 September 2017). "Polysaccharides from macroalgae: Recent advances, innovative technologies and challenges in extraction and purification". Food Research International. 99: 1011–1020. doi:10.1016/j.foodres.2016.11.016. ISSN 0963-9969. Retrieved 12 May 2023.
- ↑ Shapiro AL, Viñuela E, Maizel JV Jr (1967). "Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels". Biochem. Biophys. Res. Commun. 28 (5): 815–20. doi:10.1016/0006-291X(67)90391-9. PMID 4861258.
- ↑ Weber K, Osborn M (1969). "The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis". J Biol Chem. 244 (16): 4406–12. doi:10.1016/S0021-9258(18)94333-4. PMID 5806584.
- ↑ 19.0 19.1 Laemmli UK (1970). "Cleavage of structural proteins during the assembly of the head of bacteriophage T4". Nature. 227 (5259): 680–5. Bibcode:1970Natur.227..680L. doi:10.1038/227680a0. PMID 5432063. S2CID 3105149.
- ↑ Caprette DR. "SDS-PAGE". Experimental Biosciences. Archived from the original on 5 September 2009. Retrieved 27 September 2009.
- ↑ "2-Mercaptoethanol, Electrophoresis Grade, 98+%, Thermo Scientific Chemicals". Termofisher. Archived from the original on 1 July 2023. Retrieved 18 May 2023.
- ↑ "What is the meaning of de -gas the acrylamide gel mix?". Protocol Online. 2006. Archived from the original on 15 October 2009. Retrieved 28 September 2009.
- ↑ "SDS-PAGE". Archived from the original on 20 February 2014. Retrieved 12 September 2009.
- ↑ Rüchel R, Steere RL, Erbe EF (1978). "Transmission-electron microscopic observations of freeze-etched polyacrylamide gels". J. Chromatogr. A. 166 (2): 563–575. doi:10.1016/S0021-9673(00)95641-3.
- ↑ Schägger H, von Jagow G (1987). "Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa". Anal. Biochem. 166 (2): 368–379. doi:10.1016/0003-2697(87)90587-2. PMID 2449095.
- ↑ Ancrews D (2007). "SDS-PAGE". Andrews Lab. Archived from the original on 2 July 2017. Retrieved 27 September 2009.
- ↑ Quandt N, Stindl A, Keller U (1993). "Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis for Mr Estimations of High-Molecular-Weight Polypeptides". Anal. Biochem. 214 (2): 490–494. doi:10.1006/abio.1993.1527. PMID 8109738.
- ↑ 28.0 28.1 Brunelle, Julie L.; Green, Rachel (1 January 2014). "Chapter Thirteen - Coomassie Blue Staining". Methods in Enzymology. Academic Press. pp. 161–167.
- ↑ Green, Michael R.; Sambrook, Joseph (3 February 2020). "Staining Nucleic Acids". Cold Spring Harbor Protocols. 2020 (2): 098228. doi:10.1101/pdb.top098228. ISSN 1559-6095. Archived from the original on 1 July 2023. Retrieved 21 May 2023.
- ↑ Kumar, Gaurav (2018). "Principle and Method of Silver Staining of Proteins Separated by Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis". Methods in Molecular Biology (Clifton, N.J.). 1853: 231–236. doi:10.1007/978-1-4939-8745-0_26. ISSN 1940-6029. Archived from the original on 17 June 2022. Retrieved 21 May 2023.
- ↑ Dell'Angelica, E. C.; Bonifacino, J. S. (May 2001). "Staining proteins in gels". Current Protocols in Cell Biology. Chapter 6: Unit 6.6. doi:10.1002/0471143030.cb0606s06. ISSN 1934-2616. Archived from the original on 1 July 2023. Retrieved 11 May 2023.
- ↑ Mahmood, T; Yang, PC (September 2012). "Western blot: technique, theory, and trouble shooting". North American journal of medical sciences. 4 (9): 429–34. doi:10.4103/1947-2714.100998. PMID 23050259. Archived from the original on 8 March 2023. Retrieved 19 May 2023.
- ↑ Carlson, David P.; Watkins, Paul C.; Klevan, Leonard (22 January 1997). "Size markers for electrophoretic analysis of DNA". Archived from the original on 1 July 2023. Retrieved 23 May 2023.
- ↑ "QPNC-PAGE". www.chemeurope.com. Archived from the original on 5 December 2022. Retrieved 23 May 2023.
- ↑ Davis BJ, Ornstein L (1959). "A new high resolution electrophoresis method". Delivered at the Society for the Study of Blood at the New York Academy of Medicine.
- ↑ Raymond S, Weintraub L (1959). "Acrylamide gel as a supporting medium for zone electrophoresis". Science. 130 (3377): 711. Bibcode:1959Sci...130..711R. doi:10.1126/science.130.3377.711. PMID 14436634. S2CID 7242716.
- ↑ Rüchel R, Steere RL, Erbe EF (1978). "Transmission-electron microscopic observations of freeze-etched polyacrylamide gels". J. Chromatogr. A. 166 (2): 563–75. doi:10.1016/S0021-9673(00)95641-3.
- ↑ Bundy, D. C. (May 2001). "Autoradiography". Current Protocols in Protein Science. Chapter 10: Unit 10.11. doi:10.1002/0471140864.ps1011s10. ISSN 1934-3663. PMID 18429093. S2CID 221601750. Archived from the original on 1 July 2023. Retrieved 10 May 2023.
- ↑ "Electrophoresis for western blot" (PDF). Abcam. Archived (PDF) from the original on 22 March 2023. Retrieved 24 May 2023.
- ↑ Duchesne LG, Lam JS, MacDonald LA, et al. (1988). "Effect of pH and acrylamide concentration on the separation of lipopolysaccharides in polyacrylamide gels". Current Microbiology. 16 (4): 191–4. doi:10.1007/BF01568528. S2CID 932635.
- ↑ Brooke, Dewey; Movahed, Navid; Bothner, Brian; Brooke, Dewey; Movahed, Navid; Bothner, Brian (2015). "Universal buffers for use in biochemistry and biophysical experiments". AIMS Biophysics. 2 (3): 336–342. doi:10.3934/biophy.2015.3.336. PMC 8956001. PMID 35340547.
- ↑ Roskams, Jane; Rodgers, Linda (2002). Lab Ref: A Handbook of Recipes, Reagents, and Other Reference Tools for Use at the Bench. CSHL Press. p. 21. ISBN 978-0-87969-630-6. Archived from the original on 1 July 2023. Retrieved 17 May 2023.
- ↑ "Reproducibility with Biological Buffers". Millipore Sigma. Archived from the original on 9 February 2023. Retrieved 24 May 2023.
- ↑ Westermeier, Reiner (16 March 2016). Electrophoresis in Practice: A Guide to Methods and Applications of DNA and Protein Separations. John Wiley & Sons. p. 38. ISBN 978-3-527-69516-4. Archived from the original on 1 July 2023. Retrieved 19 May 2023.
- ↑ Ninfa, Alexander J.; Ballou, David P.; Benore, Marilee (26 May 2009). Fundamental Laboratory Approaches for Biochemistry and Biotechnology. John Wiley & Sons. p. 170. ISBN 978-0-470-08766-4. Archived from the original on 1 July 2023. Retrieved 19 May 2023.
- ↑ NARAVA, RAMANA; A, ANDHARE AISHWARYA (1 December 2020). A BEGINNER’S GUIDE TO PRACTICAL MOLECULAR ENTOMOLOGY: BASIC TECHNIQUES IN MOLECULAR BIOLOGY. American Academic Press. p. 114. ISBN 978-1-63181-504-1. Archived from the original on 1 July 2023. Retrieved 27 May 2023.
- ↑ Tareke E, Rydberg P, Eriksson S, et al. (2000). "Acrylamide: a cooking carcinogen?". Chem. Res. Toxicol. 13 (6): 517–22. doi:10.1021/tx9901938. PMID 10858325.
- ↑ LoPachin R (2004). "The changing view of acrylamide neurotoxicity". Neurotoxicology. 25 (4): 617–30. doi:10.1016/j.neuro.2004.01.004. PMID 15183015.
- ↑ Yuan-na Sun; et al. (2014). "Super Tough, Ultrastretchable, and Thermoresponsive Hydrogels with Functionalized Triblock Copolymer Micelles as Macro-Cross-Linkers". ACS Macro Letters. 3 (5): 496–500. doi:10.1021/mz500221j. PMID 35590790.
- ↑ Paez, Julieta I.; Farrukh, Aleeza; Ustahüseyin, Oya; Del Campo, Aránzazu (2018). "Biofunctionalization of Poly(acrylamide) Gels". Biomaterials for Tissue Engineering. Methods in Molecular Biology. Vol. 1758. pp. 101–114. doi:10.1007/978-1-4939-7741-3_8. ISBN 978-1-4939-7739-0. PMID 29679325.
- ↑ "Sodium dodecyl sulfate". pubchem.ncbi.nlm.nih.gov. Archived from the original on 13 February 2023. Retrieved 9 May 2023.
- ↑ Zhou, Jian-Ying; Dann, Geoffrey P.; Shi, Tujin; Wang, Lu; Gao, Xiaoli; Su, Dian; Nicora, Carrie D.; Shukla, Anil K.; Moore, Ronald J.; Liu, Tao; Camp, David G.; Smith, Richard D.; Qian, Wei-Jun (20 March 2012). "Simple sodium dodecyl sulfate-assisted sample preparation method for LC-MS-based proteomics applications". Analytical Chemistry. 84 (6): 2862–2867. doi:10.1021/ac203394r. ISSN 1520-6882. Archived from the original on 26 May 2021. Retrieved 17 May 2023.
- ↑ Singer, Michael M.; Tjeerdema, Ronald S. (1993). "Fate and Effects of the Surfactant Sodium Dodecyl Sulfate". Reviews of Environmental Contamination and Toxicology. Springer: 95–149. doi:10.1007/978-1-4613-9529-4_3. Archived from the original on 9 June 2021. Retrieved 18 May 2023.
- ↑ "SDS-PAGE". www.bionity.com. Archived from the original on 3 February 2023. Retrieved 26 May 2023.
- ↑ Aldrich, Sigma. "Urea Solution Product Information" (PDF). Archived (PDF) from the original on 7 February 2023. Retrieved 7 February 2023.
- ↑ Summer, Heike; Grämer, René; Dröge, Peter (29 October 2009). "Denaturing urea polyacrylamide gel electrophoresis (Urea PAGE)". Journal of Visualized Experiments: JoVE (32): 1485. doi:10.3791/1485. ISSN 1940-087X. Archived from the original on 24 June 2022. Retrieved 18 May 2023.
- ↑ "Ammonium Persulfate". pubchem.ncbi.nlm.nih.gov. Archived from the original on 22 November 2022. Retrieved 9 May 2023.
- ↑ "N,N,N',N'-Tetramethylethylenediamine". pubchem.ncbi.nlm.nih.gov. Archived from the original on 13 February 2023. Retrieved 9 May 2023.
- ↑ Haynes, Richard K.; Vonwiller, Simone C.; Luderer, Matthew R. (15 September 2006). "N,N,N ′, N ′-Tetramethylethylenediamine". Encyclopedia of Reagents for Organic Synthesis. John Wiley & Sons, Ltd. pp. rt064.pub2. ISBN 978-0-471-93623-7. Archived from the original on 11 February 2023. Retrieved 16 May 2023.
- ↑ "Bromophenol Blue". pubchem.ncbi.nlm.nih.gov. Archived from the original on 13 February 2023. Retrieved 9 May 2023.
- ↑ Buckingham, Lela (22 February 2019). Molecular Diagnostics: Fundamentals, Methods and Clinical Applications. F.A. Davis. p. 109. ISBN 978-0-8036-9954-0. Archived from the original on 1 July 2023. Retrieved 16 May 2023.
- ↑ Lela Buckingham and Maribeth L. Flaws (2007). Molecular Diagnostics: Fundamentals, Methods, & Clinical Applications. F.A. Davis Company. p. 91.
- ↑ Carson, Freida L; Hladik, Christa (2009). Histotechnology: A Self-Instructional Text (3 ed.). Hong Kong: American Society for Clinical Pathology Press. p. 362. ISBN 978-0-89189-581-7.
- ↑ "Coomassie Brilliant Blue R250 (C.I. 42660)". pubchem.ncbi.nlm.nih.gov. Archived from the original on 1 July 2023. Retrieved 8 May 2023.
- ↑ "Coomassie Brilliant blue R 250 (C.I. 42660)". Millapore Sigma. Archived from the original on 1 July 2023. Retrieved 23 May 2023.
- ↑ Sabnis RW (2010). Handbook of biological dyes and stains: synthesis and industrial applications. Hoboken, NJ: Wiley-Blackwell. ISBN 978-0-470-40753-0. OCLC 647922579.
- ↑ Singer VL, Lawlor TE, Yue S (1999). "Comparison of SYBR Green I nucleic acid gel stain mutagenicity and ethidium bromide mutagenicity in the Salmonella/mammalian microsome reverse mutation assay (Ames test)". Mutat. Res. 439 (1): 37–47. doi:10.1016/s1383-5718(98)00172-7. PMID 10029672.
- ↑ Ninfa AJ, Ballou DP (2004). Fundamental laboratory approaches for biochemistry and biotechnology. Hoboken, NJ: Wiley & Sons. ISBN 978-1-891786-00-6. OCLC 633862582.
- ↑ Kerenyi L, Gallyas F (1973). "Über Probleme der quantitiven Auswertung der mit physikalischer Entwicklung versilberten Agarelektrophoretogramme". Clin. Chim. Acta. 47 (3): 425–436. doi:10.1016/0009-8981(73)90276-3. PMID 4744834.
- ↑ Switzer RC 3rd, Merril CR, Shifrin S (1979). "A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels". Anal. Biochem. 98 (1): 231–7. doi:10.1016/0003-2697(79)90732-2. PMID 94518.
- ↑ Hempelmann E, Schulze M, Götze O (1984). "Free SH-groups are important for the polychromatic staining of proteins with silver nitrat". In Neuhof V (ed.). Electrophoresis '84. Weinheim: Verlag Chemie. pp. 328–30.
- ↑ Grant G (2007). "How the 1906 Nobel Prize in Physiology or Medicine was shared between Golgi and Cajal". Brain Res Rev. 55 (2): 490–8. doi:10.1016/j.brainresrev.2006.11.004. PMID 17306375. S2CID 24331507.
- ↑ Song D, Ma S, Khor SP (2002). "Gel electrophoresis-autoradiographic image analysis of radiolabeled protein drug concentration in serum for pharmacokinetic studies". Journal of Pharmacological and Toxicological Methods. 47 (1): 59–66. doi:10.1016/s1056-8719(02)00203-4. PMID 12387940.
- ↑ Yang, Ping-Chang; Mahmood, Tahrin (2012). "Western blot: Technique, theory, and trouble shooting". North American Journal of Medical Sciences. 4 (9): 429–434. doi:10.4103/1947-2714.100998. ISSN 1947-2714. PMC 3456489. PMID 23050259.
- ↑ Hnasko, Thomas S.; Hnasko, Robert M. (2015). "The Western Blot". Methods in Molecular Biology (Clifton, N.J.). 1318: 87–96. doi:10.1007/978-1-4939-2742-5_9. ISSN 1940-6029. Archived from the original on 24 June 2022. Retrieved 16 May 2023.
Further reading
- [1] Hempelmann E. SDS-Protein PAGE and Proteindetection by Silverstaining and Immunoblotting of Plasmodium falciparum proteins. in: Moll K, Ljungström J, Perlmann H, Scherf A, Wahlgren M (eds) Methods in Malaria Research, 5th edition, 2008, 263-266
External links
| Library resources about Polyacrylamide gel electrophoresis |
- SDS-PAGE: How it Works Archived 12 April 2018 at the Wayback Machine
- Demystifying SDS-PAGE Video Archived 29 July 2018 at the Wayback Machine
- Demystifying SDS-PAGE Archived 6 May 2019 at the Wayback Machine
- SDS-PAGE Calculator for customised recipes for TRIS Urea gels.
- 2-Dimensional Protein Gelelectrophoresis