Western blot

The western blot (sometimes called the protein immunoblot), or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract.[1]
Western blot technique uses three elements to achieve its task of separating a specific protein from a complex: separation by size, transfer of protein to a solid support, and marking target protein using a primary and secondary antibody to visualize.[1] A synthetic or animal-derived antibody (primary antibody) is created that recognizes and binds to a specific target protein. The electrophoresis membrane is washed in a solution containing the primary antibody, before excess antibody is washed off. A secondary antibody is added which recognizes and binds to the primary antibody. The secondary antibody is visualized through various methods such as staining, immunofluorescence, and radioactivity, allowing indirect detection of the specific target protein.[2][1]
Other techniques include dot blot analysis, quantitative dot blot, immunohistochemistry and immunocytochemistry, where antibodies are used to detect proteins in tissues and cells by immunostaining, and enzyme-linked immunosorbent assay (ELISA).[3][4][5][6]
The name western blot is a play on the Southern blot, a technique for DNA detection named after its inventor, English biologist Edwin Southern. Similarly, detection of RNA is termed as northern blot.[7] The term "western blot" was given by W. Neal Burnette in 1981,[8] although the method itself originated in 1979 in the laboratory of Harry Towbin at the Friedrich Miescher Institute in Basel, Switzerland.[9] Between 1979 and 2019 "it has been mentioned in the titles, abstracts, and keywords of more than 400,000 PubMed-listed publications" and may still be the most used protein-analytical technique.[10]
Applications
The western blot is extensively used in biochemistry for the qualitative detection of single proteins and protein-modifications (such as post-translational modifications). At least 8-9% of all protein-related publications are estimated to apply western blots.[10] It is used as a general method to identify the presence of a specific single protein within a complex mixture of proteins. A semi-quantitative estimation of a protein can be derived from the size and color intensity of a protein band on the blot membrane. The western blot is routinely used for verification of protein production after cloning.[2][11][1]It is also used in medical diagnostics, e.g., in the HIV test[12]

The confirmatory HIV test employs a western blot to detect anti-HIV antibody in a human serum sample. Proteins from known HIV-infected cells are separated and blotted on a membrane as above. Then, the serum to be tested is applied in the primary antibody incubation step; free antibody is washed away, and a secondary anti-human antibody linked to an enzyme signal is added. The stained bands then indicate the proteins to which the patient's serum contains antibody.[12] A western blot is also used as the definitive test for variant Creutzfeldt–Jakob Disease, a type of prion disease linked to the consumption of contaminated beef from cattle with bovine spongiform encephalopathy (BSE, commonly referred to as 'mad cow disease').[13]
Another application is in the diagnosis of tularemia. An evaluation of the western blot's ability to detect antibodies against F. tularensis revealed that its sensitivity is almost 100% and the specificity is 99.6%.[14]
Some forms of Lyme disease testing employ western blotting.[15] A western blot can also be used as a confirmatory test for Hepatitis B infection and HSV-2 (Herpes Type 2) infection.[16][17] In veterinary medicine, a western blot is sometimes used to confirm FIV+ status in cats.[18]
Further applications of the western blot technique include its use by the World Anti-Doping Agency (WADA). Blood doping is the misuse of certain techniques and/or substances to increase one's red blood cell mass, which allows the body to transport more oxygen to muscles and therefore increase stamina and performance. There are three widely known substances or methods used for blood doping, namely, erythropoietin (EPO), synthetic oxygen carriers and blood transfusions. Each is prohibited under WADA's List of Prohibited Substances and Methods. The western blot technique was used during the 2014 FIFA World Cup in the anti-doping campaign for that event.[19] In total, over 1000 samples were collected and analyzed by Reichel, et al.[20] in the WADA accredited Laboratory of Lausanne, Switzerland. Recent research utilizing the western blot technique showed an improved detection of EPO in blood and urine based on novel Velum SAR precast horizontal gels optimized for routine analysis.[21]
Procedure

The western blot method is composed of a gel electrophoresis to separate native proteins by 3-D structure or denatured proteins by the length of the polypeptide, followed by an electrophoretic transfer onto a membrane (mostly PVDF or nitrocellulose) and an immunostaining procedure to visualize a certain protein on the blot membrane. SDS-PAGE is generally used for the denaturing electrophoretic separation of proteins. SDS is generally used as a buffer (as well as in the gel) in order to give all proteins present a uniform negative charge, since proteins can be positively, negatively, or neutrally charged. This type of electrophoresis is known as SDS-PAGE (SDS-polyacrylamide gel electrophoresis). Prior to electrophoresis, protein samples are often boiled to denature the proteins present. This ensures that proteins are separated based on size and prevents proteases (enzymes that break down proteins) from degrading samples. Following electrophoretic separation, the proteins are transferred to a membrane (typically nitrocellulose or PVDF). The membrane is often then stained with Ponceau S in order to visualize the proteins on the blot and ensure a proper transfer occurred. Next the proteins are blocked with milk (or other blocking agents) to prevent non-specific antibody binding, and then stained with antibodies specific to the target protein.[9][22]
Lastly, the membrane will be stained with a secondary antibody that recognizes the first antibody staining, which can then be used for detection by a variety of methods. The gel electrophoresis step is included in western blot analysis to resolve the issue of the cross-reactivity of antibodies.[23][24]
Gel electrophoresis

The proteins of the sample are separated using gel electrophoresis. Separation of proteins may be by isoelectric point (pI), molecular weight, electric charge, or a combination of these factors. The nature of the separation depends on the treatment of the sample and the nature of the gel.By far the most common type of gel electrophoresis employs polyacrylamide gels and buffers loaded with sodium dodecyl sulfate (SDS). SDS-PAGE maintains polypeptides in a denatured state once they have been treated with strong reducing agents to remove secondary and tertiary structure and thus allows separation of proteins by their molecular mass. Sampled proteins become covered in the negatively charged SDS, effectively becoming anionic, and migrate towards the positively charged anode through the acrylamide mesh of the gel. Smaller proteins migrate faster through this mesh, and the proteins are thus separated according to size (usually measured in kilodaltons, kDa). The concentration of acrylamide determines the resolution of the gel – the greater the acrylamide concentration, the better the resolution of lower molecular weight proteins. The lower the acrylamide concentration, the better the resolution of higher molecular weight proteins. Proteins travel only in one dimension along the gel for most blots.[25][2][1][26]
Samples are loaded into wells in the gel. One lane is usually reserved for a marker or ladder, which is a commercially available mixture of proteins of known molecular weights, typically stained so as to form visible, coloured bands. When voltage is applied along the gel, proteins migrate through it at different speeds dependent on their size. These different rates of advancement (different electrophoretic mobilities) separate into bands within each lane. Protein bands can then be compared to the ladder bands, allowing estimation of the protein's molecular weight.[23][2][1]
Transfer

To make the proteins accessible to antibody detection, they are moved from within the gel onto a membrane, a solid support, it's an essential part of the process. There are two types of membrane: nitrocellulose (NC) or polyvinylidene difluoride (PVDF). NC membrane has high affinity for protein and its retention abilities. However, NC is brittle, and does not allow the blot to be used for re-probing, whereas PVDF membrane allows the blot to be re-probed.[1]
The most commonly used method for transferring the proteins is called electroblotting. Electroblotting uses an electric current to pull the negatively charged proteins from the gel towards the positively charged anode, and into the PVDF or NC membrane. The proteins move from within the gel onto the membrane while maintaining the organization they had within the gel. The entire stack is placed in a buffer solution which moves up the paper by capillary action, bringing the proteins with it.As a result of either transfer process, the proteins are exposed on a thin membrane layer for detection. Both varieties of membrane are chosen for their non-specific protein binding properties . Protein binding is based upon hydrophobic interactions, as well as charged interactions between the membrane and protein. [27][28][29][30]
Total protein staining
Total protein staining allows the total protein that has been successfully transferred to the membrane to be visualised, allowing the user to check the uniformity of protein transfer and to perform subsequent normalization of the target protein with the actual protein amount per lane. Normalization with the so-called "loading control" was based on immunostaining of housekeeping proteins in the classical procedure, but is heading toward total protein staining recently, due to multiple benefits.[31] At least seven different approaches for total protein staining have been described for western blot normalization: Ponceau S, stain-free techniques, Sypro Ruby, Epicocconone, Coomassie R-350, Amido Black, and Cy5.[31] In order to avoid noise of signal, total protein staining should be performed before blocking of the membrane. Nevertheless, post-antibody stainings have been described as well.[32]
Blocking
Since the membrane has been chosen for its ability to bind protein and as both antibodies and the target are proteins, steps must be taken to prevent the interactions between the membrane and the antibody used for detection of the target protein. Blocking of non-specific binding is achieved by placing the membrane in a dilute solution of protein – typically 3–5% bovine serum albumin (BSA) or non-fat dry milk (both are inexpensive) in tris-buffered saline (TBS) or I-Block, with a minute percentage (0.1%) of detergent such as Tween 20 or Triton X-100. Although non-fat dry milk is preferred due to its availability, an appropriate blocking solution is needed as not all proteins in milk are compatible with all the detection bands.[1] The protein in the dilute solution attaches to the membrane in all places where the target proteins have not attached. Thus, when the antibody is added, it cannot bind to the membrane, and therefore the only available binding site is the specific target protein. This reduces background in the final product of the western blot, leading to clearer results, and eliminates false positive[2][1]
Incubation
During the detection process the membrane is "probed" for the protein of interest with a modified antibody which is linked to a reporter enzyme; when exposed to an appropriate substrate, this enzyme drives a colorimetric reaction and produces a color. This traditionally takes place in a two-step process[2][1]
Primary antibody
The primary antibodies are generated when a host species or immune cell culture is exposed to the protein of interest . Normally, this is part of the immune response, whereas here they are harvested and used as sensitive and specific detection tools that bind the protein directly.[1][2]
After blocking, a solution of primary antibody (generally between 0.5 and 5 micrograms/mL) diluted in either PBS or TBST wash buffer is incubated with the membrane under gentle agitation for typically an hour at room temperature, or overnight at 4°C. It can also be incubated at different temperatures, with lesser temperatures being associated with more binding, both specific (to the target protein, the "signal") and non-specific ("noise"). Following incubation, the membrane is washed several times in wash buffer to remove unbound primary antibody, and thereby minimize background.[1]
Secondary antibody

After rinsing the membrane to remove unbound primary antibody, the membrane is exposed to another antibody known as the secondary antibody. Antibodies come from animal sources or hybridoma . The secondary antibody recognises and binds to the species-specific portion of the primary antibody. Therefore, an anti-mouse secondary antibody will bind to almost any mouse-sourced primary antibody, and can be referred to as an anti-species antibody (e.g. anti-mouse, anti-goat ). To allow detection of the target protein, the secondary antibody is commonly linked to biotin or a reporter enzyme such as alkaline phosphatase ,horseradish peroxidase . This means that several secondary antibodies will bind to one primary antibody and enhance the signal, allowing the detection of proteins of a much lower concentration than would be visible by SDS-PAGE alone.HRP is commonly linked to secondary antibodies to allow the detection of the target protein by chemiluminescence. The chemiluminscent substrate is cleaved by HRP, resulting in the production of luminescence. Therefore, the production of luminescence is proportional to the amount of HRP-conjugated secondary antibody, and therefore, indirectly measures the presence of the target protein. A sensitive sheet of photographic film is placed against the membrane, and exposure to the light from the reaction creates an image of the antibodies bound to the blot. A cheaper but less sensitive approach (utilizes a 4-chloronaphthol stain with 1% hydrogen peroxide[33]);where the reaction produces a dark purple stain that can be photographed without using specialized photographic film.As with the ELISPOT and ELISA procedures, the enzyme can be provided with a substrate molecule that will be converted by the enzyme to a colored reaction product that will be visible on the membrane .[2][1][34][35]
Another method of secondary antibody detection utilizes a near-infrared (NIR) fluorophore-linked antibody. The light produced from the excitation of a fluorescent dye is static, making fluorescent detection a more precise and accurate measure of the difference in the signal produced by labeled antibodies bound to proteins on a western blot. Proteins can be accurately quantified because the signal generated by the different amounts of proteins on the membranes is measured in a static state, as compared to chemiluminescence, in which light is measured in a dynamic state.[36]
A third alternative is the use of radioactive label rather than an enzyme coupled to the secondary antibody, such as labeling an antibody-binding protein like Staphylococcus Protein A or Streptavidin with a radioactive isotope of iodine.An advantage of this approach is the sensitivity of auto-radiography-based imaging, which enables highly accurate protein quantification when combined with optical software .[2][1][37]
One step
Historically, the probing process was performed in two steps because of the relative ease of producing primary and secondary antibodies in separate processes. Given the advent of high-throughput protein analysis and lower limits of detection, however, there has been interest in developing one-step probing systems that would allow the process to occur faster and with fewer consumables. This requires a probe antibody which both recognizes the protein of interest and contains a detectable label, probes which are often available for known protein tags. The primary probe is incubated with the membrane in a manner similar to that for the primary antibody in a two-step process, and then is ready for direct detection after a series of wash steps.[38][39]
Detection and visualization
After the unbound probes are washed away, the western blot is ready for detection of the probes that are labeled and bound to the protein of interest. In practical terms, not all westerns reveal protein only at one band in a membrane. Size approximations are taken by comparing the stained bands to that of the marker or ladder loaded during electrophoresis. The process is commonly repeated for a structural protein, such as actin or tubulin, that should not change between samples. The amount of target protein is normalized to the structural protein to control between groups. A superior strategy is the normalization to the total protein visualized with trichloroethanol[40][41] or epicocconone.[42]
Colorimetric detection
The colorimetric detection method depends on incubation of the western blot with a substrate that reacts with the reporter enzyme (such as peroxidase) that is bound to the secondary antibody. This converts the soluble dye into an insoluble form of a different color that precipitates next to the enzyme and thereby stains the membrane. Development of the blot is then stopped by washing away the soluble dye; protein levels are evaluated through spectrophotometry.[43][44][45]
Chemiluminescent detection

Chemiluminescent detection methods depend on incubation of the western blot with a substrate that will luminesce when exposed to the reporter on the secondary antibody. The light is then detected by CCD cameras which capture a digital image of the western blot or photographic film; the image is analysed by densitometry, which evaluates the relative amount of protein staining and quantifies the results in terms of optical density.[43][2][46]
Radioactive detection
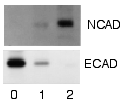
Radioactive labels do not require enzyme substrates, but rather, allow the placement of medical X-ray film directly against the western blot, which develops as it is exposed to the label and creates dark regions which correspond to the protein bands of interest. The importance of radioactive detections methods is declining due to its hazardous radiation , because it is very expensive, health and safety risks are high, and ECL (enhanced chemiluminescence) provides a useful alternative.[2][47][48]
Fluorescent detection
The fluorescently labeled probe is excited by light and the emission of the excitation is then detected by a photosensor such as a CCD camera equipped with appropriate emission filters which captures a digital image of the western blot and allows further data analysis such as molecular weight analysis and a quantitative western blot analysis. Fluorescence is considered to be one of the best methods for quantification but is less sensitive than chemiluminescence.[49][2]
2-D gel electrophoresis

2-D SDS-PAGE, as the name suggests, involves the migration of polypeptides in 2 dimensions.[50] For example, in the first dimension, polypeptides are separated according to isoelectric point, while in the second dimension, polypeptides are separated according to their molecular weight. The isoelectric point of a given protein is determined by the relative number of positively (e.g. lysine, arginine) and negatively (e.g. glutamate, aspartate) charged amino acids, with negatively charged amino acids contributing to a low isoelectric point and positively charged amino acids contributing to a high isoelectric point. Samples could also be separated first under nonreducing conditions using SDS-PAGE, and under reducing conditions in the second dimension, which breaks apart disulfide bonds that hold subunits together. SDS-PAGE might also be coupled with urea-PAGE for a 2-dimensional gel.In principle, this method allows for the separation of all cellular proteins on a single large gel. A major advantage of this method is that it often distinguishes between different isoforms of a particular protein . Proteins that have been separated can be cut out of the gel and then analysed by mass spectrometry, which identifies their molecular weight.[51][52][53][50]
Additional images
-
Western Blot Apparatus
-
Immunological Methods Western Blot
See also
- Northern blot
- Far-eastern blot
- Far-western blot
- Eastern blot
- Northwestern blot
- Fast parallel proteolysis
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Yang, Ping-Chang; Mahmood, Tahrin (2012). "Western blot: Technique, theory, and trouble shooting". North American Journal of Medical Sciences. 4 (9): 429–434. doi:10.4103/1947-2714.100998. ISSN 1947-2714. PMC 3456489. PMID 23050259.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 Gavini, Kartheek; Parameshwaran, Kodeeswaran (2021). "Western Blot". StatPearls. StatPearls Publishing. Archived from the original on 11 January 2022. Retrieved 23 December 2021.
- ↑ Farrell, Robert E. (1 January 2005). "CHAPTER 9 - Dot Blot Analysis". RNA Methodologies (Third Edition). Academic Press. pp. 179–189. ISBN 978-0-12-249696-7. Archived from the original on 13 January 2022. Retrieved 13 January 2022.
- ↑ Tian, Geng; Tang, Fangrong; Yang, Chunhua; Zhang, Wenfeng; Bergquist, Jonas; Wang, Bin; Mi, Jia; Zhang, Jiandi (19 April 2017). "Quantitative dot blot analysis (QDB), a versatile high throughput immunoblot method". Oncotarget. 8 (35): 58553–58562. doi:10.18632/oncotarget.17236. ISSN 1949-2553. Archived from the original on 13 January 2022. Retrieved 13 January 2022.
- ↑ Washington, M. KAY (1 January 2009). "CHAPTER 27 - Liver". Modern Surgical Pathology (Second Edition). W.B. Saunders. pp. 902–959. ISBN 978-1-4160-3966-2. Archived from the original on 13 January 2022. Retrieved 13 January 2022.
- ↑ Alhajj, Mandy; Farhana, Aisha (2022). "Enzyme Linked Immunosorbent Assay". StatPearls. StatPearls Publishing. Archived from the original on 25 September 2021. Retrieved 13 January 2022.
- ↑ Alwine J, Kemp D, Stark G (1977). "Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes". Proceedings of the National Academy of Sciences USA. 74 (12): 5350–5354. Bibcode:1977PNAS...74.5350A. doi:10.1073/pnas.74.12.5350. PMC 431715. PMID 414220.
- ↑ Burnette WN. (1981). "'Western blotting': electrophoretic transfer of proteins from sodium dodecyl sulfate—polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A". Analytical Biochemistry. 112 (2): 195–203. doi:10.1016/0003-2697(81)90281-5. ISSN 0003-2697. PMID 6266278.
- ↑ 9.0 9.1 Towbin H, Staehelin T, Gordon J (1979). "Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications". Proceedings of the National Academy of Sciences USA. 76 (9): 4350–54. Bibcode:1979PNAS...76.4350T. doi:10.1073/pnas.76.9.4350. PMC 411572. PMID 388439.
- ↑ 10.0 10.1 Moritz CP (2020). "40 years Western blotting: A scientific birthday toast". Journal of Proteomics. 212: 103575. doi:10.1016/j.jprot.2019.103575. PMID 31706026.
- ↑ "Western Blot". Genome.gov. Archived from the original on 10 October 2021. Retrieved 28 December 2021.
- ↑ 12.0 12.1 Sudha, T.; Lakshmi, V.; Teja, V. D. (September 2006). "Western blot profile in HIV infection". Indian Journal of Dermatology, Venereology and Leprology. 72 (5): 357–360. doi:10.4103/0378-6323.27752. ISSN 0973-3922. Archived from the original on 22 December 2021. Retrieved 14 January 2022.
- ↑ Ingrosso, Loredana; Vetrugno, Vito; Cardone, Franco; Pocchiari, Maurizio (2002-06-01). "Molecular diagnostics of transmissible spongiform encephalopathies". Trends in Molecular Medicine. 8 (6): 273–280. doi:10.1016/S1471-4914(02)02358-4. PMID 12067613.
- ↑ SCHMITT, P.; SPLETTSTÖSSER, W.; PORSCH-ÖZCÜRÜMEZ, M.; FINKE, E.-J.; GRUNOW, R. (2005-02-16). "A novel screening ELISA and a confirmatory Western blot useful for diagnosis and epidemiological studies of tularemia". Epidemiology and Infection. 133 (4): 759–766. doi:10.1017/s0950268805003742. ISSN 0950-2688. PMC 2870305. PMID 16050523.
- ↑ Artsob, Harvey (1993). "Western Blot as a confirmatory test for Lyme disease". The Canadian Journal of Infectious Diseases. 4 (2): 115–116. doi:10.1155/1993/796390. PMC 3250769. PMID 22346434.
- ↑ Castro, L De; Yoshida, C F; Gasper, A M; Gomes, S A (December 1996). "Western blot analysis of the reactivity between envelope proteins of hepatitis B viruses from Brazilian carriers and antibodies raised against recombinant hepatitis B vaccines". Acta Virol. 40 (5–6): 251–258. PMID 9171452. Archived from the original on 14 July 2021. Retrieved 6 December 2020.
- ↑ Golden, Matthew; Ashley-Morrow, Rhoda; Swenson, Paul; Hogrefe, Wayne R; Handsfield, H Hunter; Wald, Anna (December 2005). "Herpes simplex virus type 2 (HSV-2) Western blot confirmatory testing among men testing positive for HSV-2 using the focus enzyme-linked immunosorbent assay in a sexually transmitted disease clinic". Sexually Transmitted Disease. 32 (12): 771–777. doi:10.1097/01.olq.0000175377.88358.f3. PMID 16314775. S2CID 10591513. Archived from the original on 11 January 2022. Retrieved 6 December 2020.
- ↑ "FIV testing – which to use". Catwork. Archived from the original on 23 March 2021. Retrieved 6 December 2020.
- ↑ Baume, Norbert; Jan, Nicolas; Emery, Caroline; Mandanis, Béatrice; Schweizer, Carine; Giraud, Sylvain; Leuenberger, Nicolas; Marclay, François; Nicoli, Raul; Perrenoud, Laurent; Robinson, Neil; Dvorak, Jiri; Saugy, Martial (2015). "Antidoping programme and biological monitoring before and during the 2014 FIFA World Cup Brazil". British Journal of Sports Medicine. 49 (9): 614–622. doi:10.1136/bjsports-2015-094762. ISSN 0306-3674. PMC 4413745. PMID 25878079.
- ↑ Reichel C, Benetka W, Lorenc B, Thevis M (2016). "Evaluation of AMGEN clone 9G8A anti-Epo antibody for application in doping control". Drug Test Anal. 8 (11–12): 1131–1137. doi:10.1002/dta.2057. PMID 27552163.
- ↑ Schwenke, Dirk (2015). "Application Note: Improved detection of EPO in blood and urine based on novel Velum SAR precast horizontal gels optimized for routine analysis" (PDF). Archived (PDF) from the original on 2021-08-27. Retrieved 2021-12-20.
- ↑ Renart J, Reiser J, Stark GR (1979). "Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: a method for studying antibody specificity and antigen structure". Proceedings of the National Academy of Sciences USA. 76 (7): 3116–20. Bibcode:1979PNAS...76.3116R. doi:10.1073/pnas.76.7.3116. PMC 383774. PMID 91164.
- ↑ 23.0 23.1 Hnasko, Thomas S.; Hnasko, Robert M. (2015). "The Western Blot". ELISA: Methods and Protocols. Springer: 87–96. doi:10.1007/978-1-4939-2742-5_9. Archived from the original on 8 June 2018. Retrieved 27 December 2021.
- ↑ Kurien, Biji T.; Scofield, R. Hal (2015). "Western Blotting: An Introduction". Methods in molecular biology (Clifton, N.J.). 1312: 17–30. doi:10.1007/978-1-4939-2694-7_5. ISSN 1064-3745. Archived from the original on 11 January 2022. Retrieved 29 December 2021.
- ↑ Jensen, Ellen C. (2012). "The Basics of Western Blotting". The Anatomical Record. 295 (3): 369–371. doi:10.1002/ar.22424. ISSN 1932-8494. Archived from the original on 15 December 2021. Retrieved 28 December 2021.
- ↑ Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). Isolating, Cloning, and Sequencing DNA. Archived from the original on 1 January 2022. Retrieved 8 January 2022.
- ↑ Wong, Susan; Padua, Allan; Henzel, William J. (1 January 1992). "Protein and Peptide Recovery from PVDF Membranes". Techniques in Protein Chemistry III. Academic Press. pp. 3–9. ISBN 978-0-12-058756-8. Archived from the original on 16 January 2022. Retrieved 16 January 2022.
- ↑ Goldman, Aaron; Ursitti, Jeanine A.; Mozdzanowski, Jacek; Speicher, David W. (2 November 2015). "Electroblotting from Polyacrylamide Gels". Current Protocols in Protein Science. 82: 10.7.1–10.7.16. doi:10.1002/0471140864.ps1007s82. ISSN 1934-3663. Archived from the original on 23 October 2021. Retrieved 15 January 2022.
- ↑ Jr, Robert E. Farrell (31 August 2009). RNA Methodologies: Laboratory Guide for Isolation and Characterization. Academic Press. p. 249. ISBN 978-0-08-088495-0. Archived from the original on 16 January 2022. Retrieved 16 January 2022.
- ↑ Goldman, Aaron; Speicher, David W. (2 November 2015). "UNIT 10.7 Electroblotting from Polyacrylamide Gels". Current protocols in protein science / editorial board, John E. Coligan ... [et al.] 82: 10.7.1–10.7.16. doi:10.1002/0471140864.ps1007s82. ISSN 1934-3655. Archived from the original on 16 January 2022. Retrieved 16 January 2022.
- ↑ 31.0 31.1 Moritz, CP. (2017-09-20). "Tubulin or not tubulin: Heading toward total protein staining as loading control in western blots" (PDF). Proteomics. 17 (20): 1600189. doi:10.1002/pmic.201600189. PMID 28941183. S2CID 22305461. Archived (PDF) from the original on 2021-12-22. Retrieved 2021-12-20.
- ↑ Welinder, Charlotte; Ekblad, Lars (2011-03-04). "Coomassie Staining as Loading Control in Western Blot Analysis". Journal of Proteome Research. 10 (3): 1416–1419. doi:10.1021/pr1011476. ISSN 1535-3893. PMID 21186791.
- ↑ Meurant, Gerard (11 March 1996). Endocrine Methods. Elsevier. p. 214. ISBN 978-0-08-053089-5. Archived from the original on 24 January 2022. Retrieved 24 January 2022.
- ↑ Joshi, Sonali; Yu, Dihua (1 January 2017). "Chapter 8 - Immunofluorescence". Basic Science Methods for Clinical Researchers. Academic Press. pp. 135–150. ISBN 978-0-12-803077-6. Retrieved 18 January 2022.
- ↑ Clark, David P.; Pazdernik, Nanette J.; McGehee, Michelle (2 November 2018). Molecular Biology. Elsevier. p. 490. ISBN 978-0-12-813289-0. Retrieved 18 January 2022.
- ↑ Ambroz K. (2006-09-20). "Improving quantification accuracy for western blots" (PDF). Image Analysis.
{{cite journal}}: CS1 maint: url-status (link) - ↑ Dubey, R. C. (2014). Advanced Biotechnology. S. Chand Publishing. p. 361. ISBN 978-81-219-4290-4. Archived from the original on 23 January 2022. Retrieved 22 January 2022.
- ↑ "One hour Western blot" (PDF). Genscript. Archived (PDF) from the original on 13 May 2008. Retrieved 31 December 2021.
- ↑ Khan, Firdos Alam (3 September 2018). Biotechnology Fundamentals. CRC Press. p. 110. ISBN 978-1-315-36239-7. Retrieved 18 January 2022.
- ↑ Stennert, E; Arold, R (1973). "The double external auditory canal (author's transl)". HNO. 21 (10): 293–6. PMID 4769330.
- ↑ Gilda, J. E.; Gomes, A. V. (2013). "Stain-Free total protein staining is a superior loading control to β-actin for Western blots". Analytical Biochemistry. 440 (2): 186–8. doi:10.1016/j.ab.2013.05.027. PMC 3809032. PMID 23747530.
- ↑ Moritz, C. P.; Marz, S. X.; Reiss, R; Schulenborg, T; Friauf, E (2014). "Epicocconone staining: A powerful loading control for Western blots". Proteomics. 14 (2–3): 162–8. doi:10.1002/pmic.201300089. PMID 24339236. S2CID 206368546.
- ↑ 43.0 43.1 Alegria-Schaffer, Alice; Lodge, Andrew; Vattem, Krishna (2009). "Performing and optimizing Western blots with an emphasis on chemiluminescent detection". Methods in Enzymology. 463: 573–599. doi:10.1016/S0076-6879(09)63033-0. ISSN 1557-7988. Archived from the original on 11 January 2022. Retrieved 4 January 2022.
- ↑ Tomar, Rukam S. (2010). Molecular Markers and Plant Biotechnology. New India Publishing. p. 382. ISBN 978-93-80235-25-7. Archived from the original on 2022-01-19. Retrieved 2022-01-19.
- ↑ "Spectrophotometry". NIST. 13 November 2009. Archived from the original on 13 December 2020. Retrieved 19 January 2022.
- ↑ Cinquanta, Luigi; Fontana, Desré Ethel; Bizzaro, Nicola (24 June 2017). "Chemiluminescent immunoassay technology: what does it change in autoantibody detection?". Auto-Immunity Highlights. 8 (1): 9. doi:10.1007/s13317-017-0097-2. ISSN 2038-0305. Archived from the original on 20 January 2022. Retrieved 20 January 2022.
- ↑ Mruk, Dolores D; Cheng, C Yan (2011). "Enhanced chemiluminescence (ECL) for routine immunoblotting". Spermatogenesis. 1 (2): 121–122. doi:10.4161/spmg.1.2.16606. ISSN 2156-5554. Archived from the original on 28 July 2020. Retrieved 20 January 2022.
- ↑ Bakker, Onno (17 April 2013). Lumi-ImagerTM F1: Lab Protocols. Springer Science & Business Media. p. 2. ISBN 978-3-662-08431-1. Archived from the original on 21 January 2022. Retrieved 21 January 2022.
- ↑ Mathews, Suresh T.; Plaisance, Eric P.; Kim, Teayoun (2009). "Imaging Systems for Westerns: Chemiluminescence vs. Infrared Detection". Protein Blotting and Detection. Methods in Molecular Biology. Vol. 536. Humana Press. pp. 499–513. doi:10.1007/978-1-59745-542-8_51. ISBN 978-1-934115-73-2. PMID 19378087.
- ↑ 50.0 50.1 Kendrick, Nancy; Darie, Costel C.; Hoelter, Matt; Powers, Ginny; Johansen, Jon (2019). "2D SDS PAGE in Combination with Western Blotting and Mass Spectrometry Is a Robust Method for Protein Analysis with Many Applications". Advances in Experimental Medicine and Biology. 1140: 563–574. doi:10.1007/978-3-030-15950-4_33. ISSN 0065-2598. Archived from the original on 23 January 2022. Retrieved 23 January 2022.
- ↑ van der Oost, John; Walther, Jasper; Brouns, Stan JJ; van de Werken, Harmen JG; Snijders, Ambrosius PL; Wright, Phillip C; Andersson, Anders; Bernander, Rolf; de Vos, Willem M (1 January 2006). "9 Functional Genomics of the Thermo-Acidophilic Archaeon Sulfolobus solfataricus". Methods in Microbiology. Academic Press. pp. 201–231. Archived from the original on 11 January 2022. Retrieved 23 December 2021.
- ↑ O'Farrell, Patrick H. (25 May 1975). "High Resolution Two-Dimensional Electrophoresis of Proteins". The Journal of biological chemistry. 250 (10): 4007–4021. ISSN 0021-9258. Archived from the original on 25 May 2021. Retrieved 2 January 2022.
- ↑ Büyükköroğlu, Gülay; Dora, Devrim Demir; Özdemir, Filiz; Hızel, Candan (1 January 2018). "Chapter 15 - Techniques for Protein Analysis". Omics Technologies and Bio-Engineering. Academic Press. pp. 317–351. ISBN 978-0-12-804659-3. Archived from the original on 6 December 2021. Retrieved 11 January 2022.
External links
| Library resources about Western immunoblotting |
- Archived at Ghostarchive and the Wayback Machine: "Western Blotting". YouTube. Bio-Rad Laboratories. 16 October 2012.
- Archived at Ghostarchive and the Wayback Machine: "Blotting Techniques/ The Principle of Western Blotting". YouTube. Biomedical and Biological Sciences. 23 March 2017.