APPL1
Adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 (APPL1), or DCC-interacting protein 13-alpha (DIP13alpha), is a protein that in humans is encoded by the APPL1 gene.[5][6][7] APPL1 contains several key interactory domains: pleckstrin homology (PH) domain, phosphotyrosine-binding (PTB) domain and Bin–Amphiphysin–Rvs (BAR) domain.[8]
Function
APPL1 is an adaptor protein localized to a subset of Rab5-positive ("early") endosomes, where it recruits other binding partners and regulates vesicle trafficking and endosomal signalling. APPL1 is enriched at very early endosomes which are negative for EEA1, indicating that APPL1 affects the earliest stages of endosomal traffic before EEA1 takes over. This is in line with observations that APPL1 and EEA1 compete for Rab5 binding. APPL1 affects the speed of internalization of key endosomal cargo (eg. EGF receptor) which is dependent on Rab5 activation.[8]
PTB domain of APPL1 regulates many cell signalling events in specific endosomal compartments - sometimes termed the "signalling endosomes". This includes lysophosphatidic acid (LPA)-induced signaling (together with interacting protein GIPC1). Additional roles for APPL1 were pinpointed to the nucleus where APPL1 can localize once dissociated from endosomes.[8]
Mutant studies
| Mouse Mutant Alleles for Appl1 | ||
|---|---|---|
| Marker Symbol for Mouse Gene. This symbol is assigned to the genomic locus by the MGI | Appl1 | |
| Mutant Mouse Embryonic Stem Cell Clones. These are the known targeted mutations for this gene in a mouse. | Appl1tm1a(KOMP)Wtsi | |
| Example structure of targeted conditional mutant allele for this gene | ||
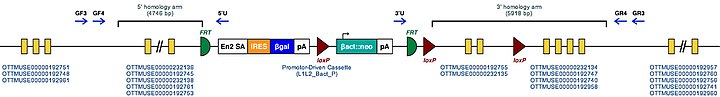
| ||
| These Mutant ES Cells can be studied directly or used to generate mice with this gene knocked out. Study of these mice can shed light on the function of Appl1:
see Knockout mouse | ||
Interactions
APPL1 has been shown to interact with Deleted in Colorectal Cancer,[9] AKT2,[5] but also Rab5, Rab21, OCRL and almost 30 other proteins.[8]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000157500 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000040760 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b Mitsuuchi Y, Johnson SW, Sonoda G, Tanno S, Golemis EA, Testa JR (September 1999). "Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2". Oncogene. 18 (35): 4891–8. doi:10.1038/sj.onc.1203080. PMID 10490823. S2CID 25245749.
- ^ Nechamen CA, Thomas RM, Dias JA (January 2007). "APPL1, APPL2, Akt2 and FOXO1a interact with FSHR in a potential signaling complex". Molecular and Cellular Endocrinology. 260–262: 93–9. doi:10.1016/j.mce.2006.08.014. PMC 1782224. PMID 17030088.
- ^ "Entrez Gene: APPL1 adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1".
- ^ a b c d Diggins NL, Webb DJ (June 2017). "APPL1 is a multifunctional endosomal signaling adaptor protein". Biochemical Society Transactions. 45 (3): 771–779. doi:10.1042/bst20160191. PMC 5844352. PMID 28620038.
- ^ Liu J, Yao F, Wu R, Morgan M, Thorburn A, Finley RL, Chen YQ (July 2002). "Mediation of the DCC apoptotic signal by DIP13 alpha". The Journal of Biological Chemistry. 277 (29): 26281–5. doi:10.1074/jbc.M204679200. PMID 12011067.
Further reading
- Nakajima D, Okazaki N, Yamakawa H, Kikuno R, Ohara O, Nagase T (June 2002). "Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones". DNA Research. 9 (3): 99–106. doi:10.1093/dnares/9.3.99. PMID 12168954.
- Nagase T, Kikuno R, Ishikawa KI, Hirosawa M, Ohara O (February 2000). "Prediction of the coding sequences of unidentified human genes. XVI. The complete sequences of 150 new cDNA clones from brain which code for large proteins in vitro". DNA Research. 7 (1): 65–73. doi:10.1093/dnares/7.1.65. PMID 10718198.
- Liu J, Yao F, Wu R, Morgan M, Thorburn A, Finley RL, Chen YQ (July 2002). "Mediation of the DCC apoptotic signal by DIP13 alpha". The Journal of Biological Chemistry. 277 (29): 26281–5. doi:10.1074/jbc.M204679200. PMID 12011067.
- Yang L, Lin HK, Altuwaijri S, Xie S, Wang L, Chang C (May 2003). "APPL suppresses androgen receptor transactivation via potentiating Akt activity". The Journal of Biological Chemistry. 278 (19): 16820–7. doi:10.1074/jbc.M213163200. PMID 12621049.
- Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M (February 2004). "APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment". Cell. 116 (3): 445–56. doi:10.1016/S0092-8674(04)00117-5. PMID 15016378. S2CID 18281503.
- Nechamen CA, Thomas RM, Cohen BD, Acevedo G, Poulikakos PI, Testa JR, Dias JA (August 2004). "Human follicle-stimulating hormone (FSH) receptor interacts with the adaptor protein APPL1 in HEK 293 cells: potential involvement of the PI3K pathway in FSH signaling". Biology of Reproduction. 71 (2): 629–36. doi:10.1095/biolreprod.103.025833. PMID 15070827.
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villén J, Li J, Cohn MA, Cantley LC, Gygi SP (August 2004). "Large-scale characterization of HeLa cell nuclear phosphoproteins". Proceedings of the National Academy of Sciences of the United States of America. 101 (33): 12130–5. Bibcode:2004PNAS..10112130B. doi:10.1073/pnas.0404720101. PMC 514446. PMID 15302935.
- Ballif BA, Villén J, Beausoleil SA, Schwartz D, Gygi SP (November 2004). "Phosphoproteomic analysis of the developing mouse brain". Molecular & Cellular Proteomics. 3 (11): 1093–101. doi:10.1074/mcp.M400085-MCP200. PMID 15345747.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Mao X, Kikani CK, Riojas RA, Langlais P, Wang L, Ramos FJ, Fang Q, Christ-Roberts CY, Hong JY, Kim RY, Liu F, Dong LQ (May 2006). "APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function". Nature Cell Biology. 8 (5): 516–23. doi:10.1038/ncb1404. PMID 16622416. S2CID 21273764.
- Cheng KK, Lam KS, Wang Y, Huang Y, Carling D, Wu D, Wong C, Xu A (May 2007). "Adiponectin-induced endothelial nitric oxide synthase activation and nitric oxide production are mediated by APPL1 in endothelial cells". Diabetes. 56 (5): 1387–94. doi:10.2337/db06-1580. PMID 17287464.
- Li J, Mao X, Dong LQ, Liu F, Tong L (May 2007). "Crystal structures of the BAR-PH and PTB domains of human APPL1". Structure. 15 (5): 525–33. doi:10.1016/j.str.2007.03.011. PMID 17502098.
- Zhu G, Chen J, Liu J, Brunzelle JS, Huang B, Wakeham N, Terzyan S, Li X, Rao Z, Li G, Zhang XC (July 2007). "Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5". The EMBO Journal. 26 (14): 3484–93. doi:10.1038/sj.emboj.7601771. PMC 1933402. PMID 17581628.
- Saito T, Jones CC, Huang S, Czech MP, Pilch PF (November 2007). "The interaction of Akt with APPL1 is required for insulin-stimulated Glut4 translocation". The Journal of Biological Chemistry. 282 (44): 32280–7. doi:10.1074/jbc.M704150200. PMID 17848569.
External links
- Human APPL1 genome location and APPL1 gene details page in the UCSC Genome Browser.







