Lentivirus
| Lentivirus | |
|---|---|
| |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Pararnavirae |
| Phylum: | Artverviricota |
| Class: | Revtraviricetes |
| Order: | Ortervirales |
| Family: | Retroviridae |
| Subfamily: | Orthoretrovirinae |
| Genus: | Lentivirus |
| Species | |
| |
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species.[2] The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.[2]
Lentiviruses can integrate a significant amount of viral complementary DNA into the DNA of the host cell and can efficiently infect nondividing cells, so they are one of the most efficient methods of gene delivery.[3][4] They can become endogenous, integrating their genome into the host germline genome, so that the virus is henceforth inherited by the host's descendants.[1]
Classification
Five serogroups of lentiviruses are recognized, reflecting the vertebrate hosts with which they are associated (primates, sheep and goats, horses, domestic cats, and cattle).[5] The primate lentiviruses are distinguished by the use of CD4 protein as a receptor and the absence of dUTPase,[6] some groups have cross-reactive gag antigens.
Morphology

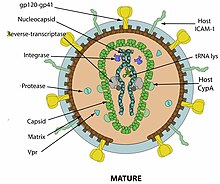
The virions are enveloped viruses 80–100 nm in diameter.[7]
They are spherical or pleomorphic, with capsid cores that mature to a cylindrical or conical shape.[7][8]
Projections of envelope make the surface appear rough, or tiny spikes (about 8 nm) may be dispersed evenly over the surface.[7]
Genome organization and replication
As with all retroviruses, lentiviruses have gag, pol and env genes, coding for viral proteins in the order: 5´-gag-pol-env-3´. Unlike other retroviruses, however, lentiviruses have two regulatory genes, tat and rev. They may also have additional accessory genes depending on the virus whose products are involved in regulation of synthesis and processing viral RNA and other replicative functions. The long terminal repeat (LTR) is about 600 nt long, of which the U3 region is 450, the R sequence 100 and the U5 region some 70 nt long.[9][7][10][11]
Retroviruses carry specific proteins within their capsids, which typically associate with the RNA genome. These proteins are typically involved in the early stages of genome replication, and include reverse transcriptase and integrase. Reverse transcriptase is the virally encoded RNA-dependent DNA polymerase. The enzyme uses the viral RNA genome as a template for the synthesis of a complementary DNA copy. Reverse transcriptase possesses RNaseH activity for destruction of the RNA-template. Integrase binds both the viral cDNA generated by reverse transcriptase and the host DNA. Integrase processes the LTR before inserting the viral genome into the host DNA. Tat acts as a trans-activator during transcription to enhance initiation and elongation. The Rev responsive element acts post-transcriptionally, regulating mRNA splicing and transport to the cytoplasm.[12][11]
Proteome

The lentiviral proteome consists of five major structural proteins and three/four non-structural proteins.[13][14]
Structural proteins listed by size:
- Gp120 surface envelope protein SU, encoded by the viral gene env. 120000 Da (daltons).[15][13]
- Gp41 transmembrane envelope protein TM, also encoded by the viral gene env. 41000 Da.[16]
- P24 capsid protein CA, encoded by the viral gene gag. 24000 Da.[17][18]
- P17 matrix protein MA, also encoded by gag. 17000 Da.[19]
- P7/P9 capsid protein NC, also encoded by gag. 7000–11000 Da.[20][13]
The envelope proteins SU and TM are glycosylated in at least some lentiviruses , if not all of them. Glycosylation seems to play a structural role in the concealment and variation of antigenic sites necessary for the host to mount an immune system response.[21][22]
Enzymes:
- Reverse transcriptase RT encoded by the pol gene.[23]
- Integrase IN also encoded by the pol gene.[10][24]
- Protease PR encoded by the pro gene [10]
- dUTPase DU encoded by the pro gene [10]
Gene regulatory proteins:[25][26]
Accessory proteins:[27]
- Nef: negative factor
- Vpr: regulatory protein
- Vif: APOBEC3 inhibitor
- Vpu/Vpx: unique to each type of HIV
Antigenic properties
Antigen determinants that possess type-specific reactivity are found on the envelope. Antigen determinants that possess type-specific reactivity and are involved in antibody mediated neutralization are found on the glycoproteins. Cross-reactivity has been found among some species of the same serotype[28][29][30]
Physicochemical and physical properties

In terms of the physicochemical and physical properties of Lentiviruses we find the following:
Classed as having class C morphology
- Nucleic acid
- Genome consists of a dimer[33]
- Virions contain one molecule of linear positive-sense single stranded RNA.[34]
- Total genome length is of one monomer ranges from 7k-13k nt [35]
- Genome sequence has terminal repeated sequences; long terminal repeats (LTR) of about 600 nt[36]
- 5' end of the genome has cap (restructure [37])
- Cap sequence of type 1 m7G5ppp5'GmpNp[38]
- There are 11 proteins
- Lipids: Virions contain 35% lipid.[40]
- Carbohydrates: Other compounds detected in the particles 3% carbohydrates.[35]
Use as gene delivery vectors

Lentivirus is primarily a research tool used to introduce a gene product into in vitro systems or animal models. Large-scale collaborative efforts are underway to use lentiviruses to block the expression of a specific gene using RNA interference technology in high-throughput formats. Conversely, lentivirus are also used to stably over-express certain genes, thus allowing researchers to examine the effect of increased gene expression in a model system.[42] [43][44][45] Though there are many challenges with Lentiviruses[46]
Another common application is to use a lentivirus to introduce a new gene into human or animal cells. For example, a model of mouse hemophilia is corrected by expressing wild-type platelet-factor VIII, the gene that is mutated in human hemophilia.[47]
Lentiviral infection has advantages over other gene-therapy methods including high-efficiency infection of dividing and non-dividing cells, long-term stable expression of a transgene, and low immunogenicity. Lentiviruses have also been successfully used for transduction of diabetic mice with the gene encoding PDGF (platelet-derived growth factor),[48] a therapy being considered for use in humans. Finally, lentiviruses have been also used to elicit an immune response against tumor antigens.[49]
These treatments, like most current gene therapy experiments, show promise but are yet to be established as safe and effective in controlled human studies. Gammaretroviral and lentiviral vectors have so far been used in more than 300 clinical trials, addressing treatment options for various diseases.[50]
Human illness

The human immunodeficiency viruses are two species of Lentivirus that infect humans. Over time, they cause acquired immunodeficiency syndrome ,[51][52] a condition in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive.[53] Without treatment, the average survival time after infection with HIV is estimated to be 9 to 11 years, depending on the HIV subtype.[54]
Signs and symptoms
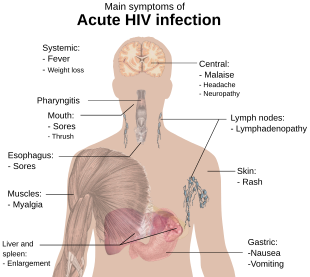
In terms of the symptoms one sees, it can start with:[55]
- Muscle pain
- Skin rash
- Headache
- Sore throat
And after some time lead to or become:[55]
- Thrush
- Cervical carcinoma in situ
- Idiopathic thrombocytopenic purpura
Or even progress to:[55]
- Opportunistic infections
- Cervical cancer (invasive)
Diagnosis
In terms of the diagnosis used to ascertain HIV we find the following:[55]
Treatment
Management of HIV/AIDS is based on a combination of medications that are termed antiretrovirals to combat the infections[55]
Epidemiology
The geographic distribution of this infection is found to be worldwide.[56]
See also
Notes
- ↑ 1.0 1.1 Kambol, R; Gatseva, A; Gifford, RJ (20 December 2022). "An endogenous lentivirus in the germline of a rodent". Retrovirology. 19 (1): 30. doi:10.1186/s12977-022-00615-2. PMID 36539757.
- ↑ 2.0 2.1 "What is Lentivirus?". News-Medical.net. 2010-05-19. Archived from the original on 2015-12-22. Retrieved 2015-11-30.
- ↑ Cockrell, Adam S.; Kafri, Tal (2007-07-01). "Gene delivery by lentivirus vectors". Molecular Biotechnology. 36 (3): 184–204. doi:10.1007/s12033-007-0010-8. ISSN 1073-6085. PMID 17873406. S2CID 25410405.
- ↑ Wang, Xiaoyin; McManus, Michael (2 October 2009). "Lentivirus Production". JoVE (Journal of Visualized Experiments) (32): e1499. doi:10.3791/1499. ISSN 1940-087X. Archived from the original on 5 March 2022. Retrieved 13 January 2024.
- ↑ Mahy, Brian W. J. (2009-02-26). The Dictionary of Virology. Academic Press. ISBN 9780080920368. Archived from the original on 2023-12-28. Retrieved 2023-12-11.
- ↑ Piguet, V.; Schwartz, O.; Le Gall, S.; Trono, D. (1999-04-01). "The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors". Immunological Reviews. 168: 51–63. doi:10.1111/j.1600-065x.1999.tb01282.x. ISSN 0105-2896. PMID 10399064. S2CID 19409388.
- ↑ 7.0 7.1 7.2 7.3 "ViralZone: Lentivirus". viralzone.expasy.org. Archived from the original on 2015-12-08. Retrieved 2015-11-30.
- ↑ Goff SP (2013). "Retroviridae". In Knipe DM, Howley PM (eds.). Fields Virology (6 ed.). Lippincott Williams & Wilkins. pp. 1424–1472. ISBN 978-1-4511-0563-6.
- ↑ Freed, E. O. (November 2001). "HIV-1 replication". Somatic Cell and Molecular Genetics. 26 (1–6): 13–33. doi:10.1023/a:1021070512287. ISSN 0740-7750. Archived from the original on 2023-01-23. Retrieved 2024-01-03.
- ↑ 10.0 10.1 10.2 10.3 Petropoulos, C. (1997). "Retroviral Taxonomy, Protein Structures, Sequences, and Genetic Maps". Retroviruses. Cold Spring Harbor Laboratory Press. Archived from the original on 29 June 2023. Retrieved 12 January 2024.
- ↑ 11.0 11.1 Tang, Hengli; Kuhen, Kelli L; Wong-Staal, Flossie (December 1999). "Lentivirus Replication and Regulation". Annual Review of Genetics. 33 (1): 133–170. doi:10.1146/annurev.genet.33.1.133. Archived from the original on 2024-01-15. Retrieved 2024-01-12.
- ↑ Buchschacher, Gary L. (2003-01-31). Lentiviral Vector Systems for Gene Transfer. Springer US. ISBN 978-0-306-47702-7. Archived from the original on 2023-12-28. Retrieved 2023-12-11.
- ↑ 13.0 13.1 13.2 13.3 "High-precision lentivirus titer determination and protein profiling" (PDF). SCIEX. Archived (PDF) from the original on 12 August 2023. Retrieved 1 January 2024.
- ↑ 14.0 14.1 Mushahwar, Isa K. (1 January 2006). "Human Immunodeficiency Viruses: Molecular Virology, Pathogenesis, Diagnosis and Treatment". Perspectives in Medical Virology. Elsevier. 13: 75–87.
- ↑ Sodroski, Joseph; Patarca, Roberto; Perkins, Dennis; Briggs, Debra; Tun-Hou, Lee; Essex, Myron; Coligan, John; Wong-Staal, Flossie; Gallo, Robert C. (1984). "Sequence of the Envelope Glycoprotein Gene of Type II Human T Lymphotropic Virus". Science. 225 (4660): 421–424. Bibcode:1984Sci...225..421S. doi:10.1126/science.6204380. PMID 6204380. Archived from the original on 2023-11-03. Retrieved 2024-01-07.
- ↑ Zwick, Michael B.; Saphire, Erica O.; Burton, Dennis R. (February 2004). "gp41: HIV's shy protein". Nature Medicine. 10 (2): 133–134. doi:10.1038/nm0204-133. ISSN 1546-170X. Archived from the original on 2022-10-20. Retrieved 2024-01-07.
- ↑ Jones, Ian M.; Morikawa, Yuko (April 1998). "The molecular basis of HIV capsid assembly". Reviews in Medical Virology. 8 (2): 87–95. doi:10.1002/(SICI)1099-1654(199804/06)8:2<87::AID-RMV220>3.0.CO;2-B. ISSN 1052-9276. Archived from the original on 2020-06-07. Retrieved 2024-01-08.
- ↑ Teow, Sin-Yeang. "Cell-Penetrating Antibodies For Targeting HIV-1 p24 Capsid Protein". Archived from the original on 2 August 2022. Retrieved 10 January 2024.
{{cite journal}}: Cite journal requires|journal=(help) - ↑ Fiorentini, Simona; Marini, Elena; Caracciolo, Sonia; Caruso, Arnaldo (January 2006). "Functions of the HIV-1 matrix protein p17". The New Microbiologica. 29 (1): 1–10. ISSN 1121-7138. Archived from the original on 2022-06-15. Retrieved 2024-01-07.
- ↑ Bell, Neil M.; Lever, Andrew M.L. (March 2013). "HIV Gag polyprotein: processing and early viral particle assembly" (PDF). Trends in Microbiology. 21 (3): 136–144. doi:10.1016/j.tim.2012.11.006. Archived (PDF) from the original on 2022-09-06. Retrieved 2024-01-07.
- ↑ Yolitz, Jason; Schwing, Catherine; Chang, Julia; Van Ryk, Donald; Nawaz, Fatima; Wei, Danlan; Cicala, Claudia; Arthos, James; Fauci, Anthony S. (6 March 2018). "Signal peptide of HIV envelope protein impacts glycosylation and antigenicity of gp120". Proceedings of the National Academy of Sciences. 115 (10): 2443–2448. doi:10.1073/pnas.1722627115. ISSN 0027-8424. Archived from the original on 31 December 2023. Retrieved 30 December 2023.
- ↑ Behrens, Anna-Janina; Crispin, Max (June 2017). "Structural principles controlling HIV envelope glycosylation". Current Opinion in Structural Biology. 44: 125–133. doi:10.1016/j.sbi.2017.03.008. ISSN 1879-033X. Archived from the original on 2023-05-09. Retrieved 2024-01-03.
- ↑ Buchschacher, Gary L.; Wong-Staal, Flossie (15 April 2000). "Development of lentiviral vectors for gene therapy for human diseases". Blood. 95 (8): 2499–2504. doi:10.1182/blood.V95.8.2499. Archived from the original on 20 October 2023. Retrieved 12 January 2024.
- ↑ "The Lentivirus System – An Introduction". info.abmgood.com. Archived from the original on 25 September 2023. Retrieved 12 January 2024.
- ↑ Debaisieux, Solène; Rayne, Fabienne; Yezid, Hocine; Beaumelle, Bruno (March 2012). "The Ins and Outs of HIV ‐1 Tat". Traffic. 13 (3): 355–363. doi:10.1111/j.1600-0854.2011.01286.x. ISSN 1398-9219. Archived from the original on 2022-08-06. Retrieved 2024-01-04.
- ↑ Pollard, V. W.; Malim, M. H. (1998). "The HIV-1 Rev protein". Annual Review of Microbiology. 52: 491–532. doi:10.1146/annurev.micro.52.1.491. ISSN 0066-4227. Archived from the original on 2023-12-19. Retrieved 2024-01-04.
- ↑ Milani, Alireza; Baesi, Kazem; Agi, Elnaz; Marouf, Ghazal; Ahmadi, Maryam; Bolhassani, Azam (2021). "HIV-1 Accessory Proteins: Which one is Potentially Effective in Diagnosis and Vaccine Development?". Protein and Peptide Letters. 28 (6): 687–698. doi:10.2174/0929866528999201231213610. ISSN 1875-5305. Archived from the original on 15 January 2024. Retrieved 11 January 2024.
- ↑ Clements, Janice E.; Gdovin, Susan L.; Montelaro, Ronald C.; Narayan, Opendra (April 1988). "Antigenic Variation in Lentiviral Diseases". Annual Review of Immunology. 6 (1): 139–159. doi:10.1146/annurev.iy.06.040188.001035. Archived from the original on 2024-01-03. Retrieved 2023-12-31.
- ↑ Yang, Lili; Bailey, Leslie; Baltimore, David; Wang, Pin (1 August 2006). "Targeting lentiviral vectors to specific cell types in vivo". Proceedings of the National Academy of Sciences of the United States of America. 103 (31): 11479–11484. doi:10.1073/pnas.0604993103. ISSN 0027-8424. Archived from the original on 15 January 2024. Retrieved 14 January 2024.
- ↑ VandeWoude, Sue; Troyer, Jennifer; Poss, Mary (15 March 2010). "Restrictions to cross-species transmission of lentiviral infection gleaned from studies of FIV". Veterinary Immunology and Immunopathology. 134 (1–2): 25–32. doi:10.1016/j.vetimm.2009.10.005. ISSN 1873-2534. Archived from the original on 15 January 2024. Retrieved 14 January 2024.
- ↑ Biasin, Mara; Bianco, Andrea; Pareschi, Giovanni; Cavalleri, Adalberto; Cavatorta, Claudia; Fenizia, Claudio; Galli, Paola; Lessio, Luigi; Lualdi, Manuela; Tombetti, Enrico; Ambrosi, Alessandro; Redaelli, Edoardo Maria Alberto; Saulle, Irma; Trabattoni, Daria; Zanutta, Alessio; Clerici, Mario (18 March 2021). "UV-C irradiation is highly effective in inactivating SARS-CoV-2 replication". Scientific Reports. 11 (1). doi:10.1038/s41598-021-85425-w.
- ↑ Nakagomi, Osamu. "Fundamentals of Ultracentrifugal Virus Purification" (PDF). beckmancoulter. Archived (PDF) from the original on 7 March 2023. Retrieved 1 January 2024.
- ↑ Tran, Thao; Liu, Yuanyuan; Marchant, Jan; Monti, Sarah; Seu, Michelle; Zaki, Jessica; Yang, Ae Lim; Bohn, Jennifer; Ramakrishnan, Venkateswaran; Singh, Rashmi; Hernandez, Mateo; Vega, Alexander; Summers, Michael F. (29 September 2015). "Conserved determinants of lentiviral genome dimerization". Retrovirology. 12 (1): 83. doi:10.1186/s12977-015-0209-x. ISSN 1742-4690. Archived from the original on 11 October 2022. Retrieved 15 January 2024.
- ↑ Duvergé, Alexis; Negroni, Matteo (16 November 2020). "Pseudotyping Lentiviral Vectors: When the Clothes Make the Virus". Viruses. 12 (11): 1311. doi:10.3390/v12111311. ISSN 1999-4915. Archived from the original on 15 January 2024. Retrieved 15 January 2024.
- ↑ 35.0 35.1 Viruses, International Committee on Taxonomy of; King, Andrew MQ (25 October 2011). Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier. p. 478. ISBN 978-0-12-384684-6. Archived from the original on 16 January 2024. Retrieved 16 January 2024.
- ↑ Schambach, Axel; Zychlinski, Daniela; Ehrnstroem, Birgitta; Baum, Christopher (February 2013). "Biosafety Features of Lentiviral Vectors". Human Gene Therapy. 24 (2): 132–142. doi:10.1089/hum.2012.229. ISSN 1043-0342. Archived from the original on 15 January 2024. Retrieved 15 January 2024.
- ↑ Counsell, John R.; De Brabandere, Guillaume; Karda, Rajvinder; Moore, Marc; Greco, Antonio; Bray, Alysha; Diaz, Juan Antinao; Perocheau, Dany P.; Mock, Ulrike; Waddington, Simon N. (15 December 2020). "Re-structuring lentiviral vectors to express genomic RNA via cap-dependent translation". Molecular Therapy. Methods & Clinical Development. 20: 357–365. doi:10.1016/j.omtm.2020.12.005. ISSN 2329-0501. Archived from the original on 15 January 2024. Retrieved 15 January 2024.
- ↑ Lu, Pu-Xuan; Zhou, Bo-Ping (26 November 2015). Diagnostic Imaging of Emerging Infectious Diseases. Springer. pp. 77–78. ISBN 978-94-017-7363-8. Archived from the original on 15 January 2024. Retrieved 15 January 2024.
- ↑ "Lentiviral Gene Delivery for Mammalian Expression—Getting Started - US". www.thermofisher.com. Archived from the original on 22 October 2015. Retrieved 15 January 2024.
- ↑ "Family - Retroviridae". Virus Taxonomy. Elsevier. 1 January 2012. pp. 477–495. ISBN 978-0-12-384684-6. Retrieved 16 January 2024.
- ↑ Milone, Michael C.; O’Doherty, Una (2018-03-22). "Clinical use of lentiviral vectors". Leukemia. Springer Science and Business Media LLC. 32 (7): 1529–1541. doi:10.1038/s41375-018-0106-0. ISSN 0887-6924.
- ↑ "shRNA – short hairpin RNA". Archived from the original on 2008-10-02. Retrieved 2023-12-11.
- ↑ "Casgevy | European Medicines Agency". www.ema.europa.eu. Archived from the original on 2023-12-19. Retrieved 2023-12-29.
- ↑ Dong, Wendy; Kantor, Boris (July 2021). "Lentiviral Vectors for Delivery of Gene-Editing Systems Based on CRISPR/Cas: Current State and Perspectives". Viruses. 13 (7): 1288. doi:10.3390/v13071288. ISSN 1999-4915. Archived from the original on 2022-10-13. Retrieved 2024-01-05.
- ↑ Gutierrez-Guerrero, Alejandra; Cosset, François-Loïc; Verhoeyen, Els (September 2020). "Lentiviral Vector Pseudotypes: Precious Tools to Improve Gene Modification of Hematopoietic Cells for Research and Gene Therapy". Viruses. 12 (9): 1016. doi:10.3390/v12091016. ISSN 1999-4915. Archived from the original on 2022-11-02. Retrieved 2024-01-05.
- ↑ Martínez-Molina, Eduardo; Chocarro-Wrona, Carlos; Martínez-Moreno, Daniel; Marchal, Juan A.; Boulaiz, Houria (3 November 2020). "Large-Scale Production of Lentiviral Vectors: Current Perspectives and Challenges". Pharmaceutics. 12 (11): 1051. doi:10.3390/pharmaceutics12111051. ISSN 1999-4923. Archived from the original on 10 January 2024. Retrieved 9 January 2024.
- ↑ Shi Q, Wilcox DA, Fahs SA, et al. (February 2007). "Lentivirus-mediated platelet-derived factor VIII gene therapy in murine haemophilia A". J. Thromb. Haemost. 5 (2): 352–61. doi:10.1111/j.1538-7836.2007.02346.x. PMID 17269937.
- ↑ Lee JA, Conejero JA, Mason JM, et al. (August 2005). "Lentiviral transfection with the PDGF-B gene improves diabetic wound healing". Plast. Reconstr. Surg. 116 (2): 532–8. doi:10.1097/01.prs.0000172892.78964.49. PMID 16079687. S2CID 8628077.
- ↑ Casado, Javier Garcia; Janda, Jozef; Wei, Joe; Chapatte, Laurence; Colombetti, Sara; Alves, Pedro; Ritter, Gerd; Ayyoub, Maha; Valmori, Danila; Chen, Weisan; Lévy, Frédéric (2008). "Lentivector immunization induces tumor antigen-specific B and T cell responses in vivo". European Journal of Immunology. 38 (7): 1867–1876. doi:10.1002/eji.200737923. ISSN 1521-4141. PMID 18546142.
- ↑ Kurth, R; Bannert, N, eds. (2010). Retroviruses: Molecular Biology, Genomics and Pathogenesis. Caister Academic Press. ISBN 978-1-904455-55-4.
- ↑ Weiss RA (May 1993). "How does HIV cause AIDS?". Science. 260 (5112): 1273–9. Bibcode:1993Sci...260.1273W. doi:10.1126/science.8493571. PMID 8493571.
- ↑ Douek DC, Roederer M, Koup RA (2009). "Emerging Concepts in the Immunopathogenesis of AIDS". Annual Review of Medicine. 60: 471–84. doi:10.1146/annurev.med.60.041807.123549. PMC 2716400. PMID 18947296.
- ↑ Powell MK, Benková K, Selinger P, Dogoši M, Kinkorová Luňáčková I, Koutníková H, Laštíková J, Roubíčková A, Špůrková Z, Laclová L, Eis V, Šach J, Heneberg P (2016). "Opportunistic Infections in HIV-Infected Patients Differ Strongly in Frequencies and Spectra between Patients with Low CD4+ Cell Counts Examined Postmortem and Compensated Patients Examined Antemortem Irrespective of the HAART Era". PLOS ONE. 11 (9): e0162704. Bibcode:2016PLoSO..1162704P. doi:10.1371/journal.pone.0162704. PMC 5017746. PMID 27611681.
- ↑ UNAIDS; WHO (December 2007). "2007 AIDS epidemic update" (PDF). p. 16. Archived (PDF) from the original on 2019-10-17. Retrieved 2023-12-27.
- ↑ 55.0 55.1 55.2 55.3 55.4 Justiz Vaillant, Angel A.; Gulick, Peter G. (2023). "HIV and AIDS Syndrome". StatPearls. StatPearls Publishing. PMID 30521281. Archived from the original on 2023-09-19. Retrieved 2023-12-28.
- ↑ "Global HIV & AIDS statistics — Fact sheet". www.unaids.org. Archived from the original on 4 December 2019. Retrieved 1 January 2024.
References
- Ryan KJ, Ray CG, eds. (2004). Sherris Medical Microbiology: An Introduction to Infectious Diseases (4th ed.). New York: McGraw Hill. ISBN 978-0-8385-8529-0.
- Desport, M, ed. (2010). Lentiviruses and Macrophages: Molecular and Cellular Interactions. Caister Academic Press. ISBN 978-1-904455-60-8.
- Knipe DM, Howley PM, eds. (2013). Fields Virology (6 ed.). Lippincott Williams & Wilkins. ISBN 978-1-4511-0563-6.
Further reading
- "Lentiviruses In Ungulates. I. General Features, History And Prevalence" (PDF). Bulgarian Journal of Veterinary Medicine. 9 (3): 175–181. 2006. Archived (PDF) from the original on 2007-09-27. Retrieved 2023-12-11.
- Tim Ravenscroft (2008-06-15). "Are Lentiviral Vectors on Cusp of Breakout?". Genetic Engineering & Biotechnology News. Mary Ann Liebert, Inc. pp. 54–55. Archived from the original on 2008-12-11. Retrieved 2008-07-06.
(subtitle) Rapidly emerging technology has potential to treat hemophilia, AIDS, and Cancer
- Katzourakis, Aris; Tristem, Michael; Pybus, Oliver G.; Gifford, Robert J. (10 April 2007). "Discovery and analysis of the first endogenous lentivirus". Proceedings of the National Academy of Sciences of the United States of America. 104 (15): 6261–6265. doi:10.1073/pnas.0700471104. ISSN 0027-8424. Archived from the original on 4 December 2021. Retrieved 13 January 2024.
External links
- Viralzone: Lentivirus Archived 2010-06-13 at the Wayback Machine
- ICTV taxonomy of Lentivirus Archived 2006-04-18 at the Wayback Machine