Video:Severe acute respiratory syndrome coronavirus 2
| Severe acute respiratory syndrome coronavirus 2 (Tutorial) | |
|---|---|
| On Commons | |
| Steps for video creation | |
| Step 1 | Preview my changes (10 sec) |
| Step 2 | Upload to Commons (10 min) |
Definition
Severe acute respiratory syndrome coronavirus 2[1] is a strain of coronavirus that causes COVID-19 , the illness responsible for the COVID-19 pandemic.[2] First identified in the city of Wuhan, Hubei, China, the World Health Organization declared the outbreak a pandemic in March 2020.[3][4]

Transmission
Human to human transmission of SARS‑CoV‑2 was confirmed on 20 January 2020 during the COVID-19 pandemic.[5][6] Transmission was initially assumed to occur primarily via respiratory droplets from coughs and sneezes within a range of about 1.8 meters.[7][8] Laser light scattering experiments suggest that speaking is an additional mode of transmission.[9]

Taxonomy
SARS‑CoV‑2 belongs to the broad family of viruses known as coronaviruses.[10] It is a positive-sense single-stranded RNA virus, with a single linear RNA segment. Coronaviruses infect humans, other mammals, including livestock and companion animals, and avian species.[11]Like the SARS-related coronavirus implicated in the 2003 SARS outbreak, SARS‑CoV‑2 is a member of the subgenus Sarbecovirus beta-CoV lineage B.[12][13]

Variants 1
The World Health Organization has currently declared five variants of concern, which are as follows,[14]Alpha Lineage B.1.1.7 emerged in the United Kingdom in September 2020, with evidence of increased transmissibility and virulence. Notable mutations include N501Y and P681H.Beta Lineage B.1.351 emerged in South Africa in May 2020, with evidence of increased transmissibility and changes to antigenicity.[14]

Variants 2
Gamma Lineage P.1 emerged in Brazil in November 2020, also with evidence of increased transmissibility and virulence, alongside changes to antigenicity. Delta Lineage B.1.617.2 emerged in India in October 2020. There is also evidence of increased transmissibility and changes to antigenicity.Omicron Lineage B.1.1.529 emerged in Botswana in November 2021.[14]

Reservoir
Bats are considered the most likely natural reservoir of SARS‑CoV‑2.[15][16] Differences between the bat coronavirus and SARS‑CoV‑2 suggest that humans may have been infected via an intermediate host,[17] although the source of introduction into humans remains unknown.[18][19]

Virology 1
Each SARS-CoV-2 virion is 60 to 140 nanometers in diameter,[20][21] its mass within the global human populace has been estimated as being between 0.1 and 1.0 kg.[22] Like other coronaviruses, SARS-CoV-2 has four structural proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope.[23]

Virology 2
SARS-CoV-2 has a linear, positive-sense, single-stranded RNA genome about 30,000 bases long.[11] Its genome has a bias against cytosine and guanine nucleotides, like other coronaviruses.[24] The genome has the highest composition of U, 32.2 percent, followed by A, 29.9 percent, and a similar composition of G, 19.6 percent and C, 18.3 percent.[25]
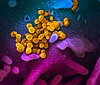
Virology 3
Virus infections start when viral particles bind to host surface cellular receptors.[26] Protein modeling experiments on the spike protein of the virus soon suggested that SARS‑CoV‑2 has sufficient affinity to the receptor angiotensin converting enzyme 2 on human cells to use them as a mechanism of cell entry.[27]

References
- ↑ Zimmer, Carl (26 February 2021). "Opinion | The Secret Life of a Coronavirus". The New York Times. Retrieved 15 February 2022.
- ↑ "WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020". www.who.int. Retrieved 15 February 2022.
- ↑ "Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV)". www.who.int. Retrieved 15 February 2022.
- ↑ Chan, Jasper Fuk-Woo; Yuan, Shuofeng; Kok, Kin-Hang; To, Kelvin Kai-Wang; Chu, Hin; Yang, Jin; Xing, Fanfan; Liu, Jieling; Yip, Cyril Chik-Yan; Poon, Rosana Wing-Shan; Tsoi, Hoi-Wah; Lo, Simon Kam-Fai; Chan, Kwok-Hung; Poon, Vincent Kwok-Man; Chan, Wan-Mui; Ip, Jonathan Daniel; Cai, Jian-Piao; Cheng, Vincent Chi-Chung; Chen, Honglin; Hui, Christopher Kim-Ming; Yuen, Kwok-Yung (2020). "A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster". Lancet (London, England). 395 (10223): 514–523. doi:10.1016/S0140-6736(20)30154-9. ISSN 0140-6736. Retrieved 16 February 2022.
- ↑ Li, Jin-Yan; You, Zhi; Wang, Qiong; Zhou, Zhi-Jian; Qiu, Ye; Luo, Rui; Ge, Xing-Yi (2020). "The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future". Microbes and Infection. 22 (2): 80–85. doi:10.1016/j.micinf.2020.02.002. ISSN 1286-4579. Retrieved 16 February 2022.
- ↑ "Transmission of Novel Coronavirus (2019-nCoV) | CDC". web.archive.org. cdc. 28 January 2020. Retrieved 16 February 2022.
- ↑ "How does coronavirus spread?". NBC News. Retrieved 16 February 2022.
- ↑ Anfinrud, Philip; Stadnytskyi, Valentyn; Bax, Christina E.; Bax, Adriaan (15 April 2020). "Visualizing Speech-Generated Oral Fluid Droplets with Laser Light Scattering". The New England Journal of Medicine: NEJMc2007800. doi:10.1056/NEJMc2007800. ISSN 0028-4793. Retrieved 16 February 2022.
- ↑ Fox D (January 2020). "What you need to know about the novel coronavirus". Nature. doi:10.1038/d41586-020-00209-y. PMID 33483684. S2CID 213064026.
- ↑ 11.0 11.1 V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V (March 2021). "Coronavirus biology and replication: implications for SARS-CoV-2". Nature Reviews. Microbiology. 19 (3): 155–170. doi:10.1038/s41579-020-00468-6. PMC 7592455. PMID 33116300.
- ↑ "auspice". nextstrain.org. Retrieved 20 February 2022.
- ↑ Wong, Antonio C. P.; Li, Xin; Lau, Susanna K. P.; Woo, Patrick C. Y. (20 February 2019). "Global Epidemiology of Bat Coronaviruses". Viruses. 11 (2): 174. doi:10.3390/v11020174. ISSN 1999-4915. Retrieved 20 February 2022.
- ↑ 14.0 14.1 14.2 World Health Organization (27 Nov 2021). "Tracking SARS-CoV-2 variants". World Health Organization. Archived from the original on 6 June 2021. Retrieved 28 Nov 2021.
- ↑ "Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19)" (PDF). World Health Organization. Retrieved 17 February 2022.
- ↑ Lu, Roujian; Zhao, Xiang; Li, Juan; Niu, Peihua; Yang, Bo; Wu, Honglong; Wang, Wenling; Song, Hao; Huang, Baoying; Zhu, Na; Bi, Yuhai; Ma, Xuejun; Zhan, Faxian; Wang, Liang; Hu, Tao; Zhou, Hong; Hu, Zhenhong; Zhou, Weimin; Zhao, Li; Chen, Jing; Meng, Yao; Wang, Ji; Lin, Yang; Yuan, Jianying; Xie, Zhihao; Ma, Jinmin; Liu, William J; Wang, Dayan; Xu, Wenbo; Holmes, Edward C; Gao, George F; Wu, Guizhen; Chen, Weijun; Shi, Weifeng; Tan, Wenjie (2020). "Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding". Lancet (London, England). 395 (10224): 565–574. doi:10.1016/S0140-6736(20)30251-8. ISSN 0140-6736. Retrieved 17 February 2022.
- ↑ Cyranoski, David (26 February 2020). "Mystery deepens over animal source of coronavirus". Nature. 579 (7797): 18–19. doi:10.1038/d41586-020-00548-w. Retrieved 17 February 2022.
- ↑ O'Keeffe J, Freeman S, Nicol A (21 March 2021). The Basics of SARS-CoV-2 Transmission. Vancouver, BC: National Collaborating Centre for Environmental Health (NCCEH). ISBN 978-1-988234-54-0. Archived from the original on 12 May 2021. Retrieved 12 May 2021.
- ↑ Holmes EC, Goldstein SA, Rasmussen AL, Robertson DL, Crits-Christoph A, Wertheim JO, et al. (August 2021). "The Origins of SARS-CoV-2: A Critical Review". Cell. 184 (19): 4848–4856. doi:10.1016/j.cell.2021.08.017. PMC 8373617. PMID 34480864.
- ↑ Zhu, Na; Zhang, Dingyu; Wang, Wenling; Li, Xingwang; Yang, Bo; Song, Jingdong; Zhao, Xiang; Huang, Baoying; Shi, Weifeng; Lu, Roujian; Niu, Peihua; Zhan, Faxian; Ma, Xuejun; Wang, Dayan; Xu, Wenbo; Wu, Guizhen; Gao, George F.; Tan, Wenjie (20 February 2020). "A Novel Coronavirus from Patients with Pneumonia in China, 2019". The New England Journal of Medicine. 382 (8): 727–733. doi:10.1056/NEJMoa2001017. ISSN 0028-4793. Retrieved 20 February 2022.
- ↑ Chen, Nanshan; Zhou, Min; Dong, Xuan; Qu, Jieming; Gong, Fengyun; Han, Yang; Qiu, Yang; Wang, Jingli; Liu, Ying; Wei, Yuan; Xia, Jia'an; Yu, Ting; Zhang, Xinxin; Zhang, Li (2020). "Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study". Lancet (London, England). 395 (10223): 507–513. doi:10.1016/S0140-6736(20)30211-7. ISSN 0140-6736. Retrieved 18 February 2022.
- ↑ Sender R, Bar-On YM, Gleizer S, Bernsthein B, Flamholz A, Phillips R, Milo R (April 2021). "The total number and mass of SARS-CoV-2 virions". medRxiv: 2020.11.16.20232009. doi:10.1101/2020.11.16.20232009. PMC 7685332. PMID 33236021.
- ↑ Wu, Canrong; Liu, Yang; Yang, Yueying; Zhang, Peng; Zhong, Wu; Wang, Yali; Wang, Qiqi; Xu, Yang; Li, Mingxue; Li, Xingzhou; Zheng, Mengzhu; Chen, Lixia; Li, Hua (2020). "Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods". Acta Pharmaceutica Sinica. B. 10 (5): 766–788. doi:10.1016/j.apsb.2020.02.008. ISSN 2211-3835. Retrieved 18 February 2022.
- ↑ Kandeel M, Ibrahim A, Fayez M, Al-Nazawi M (June 2020). "From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes". Journal of Medical Virology. 92 (6): 660–666. doi:10.1002/jmv.25754. PMC 7228358. PMID 32159237.
- ↑ Hou W (September 2020). "Characterization of codon usage pattern in SARS-CoV-2". Virology Journal. 17 (1): 138. doi:10.1186/s12985-020-01395-x. PMC 7487440. PMID 32928234.
- ↑ Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. (May 2020). "Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2". Cell. 181 (4): 894–904.e9. doi:10.1016/j.cell.2020.03.045. PMC 7144619. PMID 32275855.
- ↑ Xu, Xintian; Chen, Ping; Wang, Jingfang; Feng, Jiannan; Zhou, Hui; Li, Xuan; Zhong, Wu; Hao, Pei (2020). "Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission". Science China. Life Sciences. 63 (3): 457–460. doi:10.1007/s11427-020-1637-5. ISSN 1674-7305. Retrieved 19 February 2022.