Serial analysis of gene expression
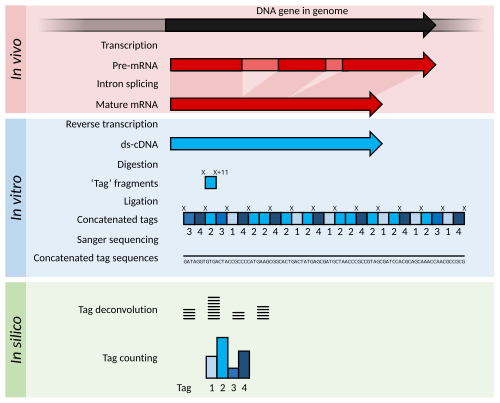
Serial Analysis of Gene Expression (SAGE) is a transcriptomic technique used by molecular biologists to produce a snapshot of the messenger RNA population in a sample of interest in the form of small tags that correspond to fragments of those transcripts. Several variants have been developed since, most notably a more robust version, LongSAGE,[2] RL-SAGE[3] and the most recent SuperSAGE.[4] Many of these have improved the technique with the capture of longer tags, enabling more confident identification of a source gene.
Overview
Briefly, SAGE experiments proceed as follows:
- The mRNA of an input sample (e.g. a tumour) is isolated and a reverse transcriptase and biotinylated primers are used to synthesize cDNA from mRNA.
- The cDNA is bound to Streptavidin beads via interaction with the biotin attached to the primers, and is then cleaved using a restriction endonuclease called an anchoring enzyme (AE). The location of the cleavage site and thus the length of the remaining cDNA bound to the bead will vary for each individual cDNA (mRNA).
- The cleaved cDNA downstream from the cleavage site is then discarded, and the remaining immobile cDNA fragments upstream from cleavage sites are divided in half and exposed to one of two adaptor oligonucleotides (A or B) containing several components in the following order upstream from the attachment site: 1) Sticky ends with the AE cut site to allow for attachment to cleaved cDNA; 2) A recognition site for a restriction endonuclease known as the tagging enzyme (TE), which cuts about 15 nucleotides downstream of its recognition site (within the original cDNA/mRNA sequence); 3) A short primer sequence unique to either adaptor A or B, which will later be used for further amplification via PCR.
- After adaptor ligation, cDNA are cleaved using TE to remove them from the beads, leaving only a short "tag" of about 11 nucleotides of original cDNA (15 nucleotides minus the 4 corresponding to the AE recognition site).
- The cleaved cDNA tags are then repaired with DNA polymerase to produce blunt end cDNA fragments.
- These cDNA tag fragments (with adaptor primers and AE and TE recognition sites attached) are ligated, sandwiching the two tag sequences together, and flanking adaptors A and B at either end. These new constructs, called ditags, are then PCR amplified using anchor A and B specific primers.
- The ditags are then cleaved using the original AE, and allowed to link together with other ditags, which will be ligated to create a cDNA concatemer with each ditag being separated by the AE recognition site.
- These concatemers are then transformed into bacteria for amplification through bacterial replication.
- The cDNA concatemers can then be isolated and sequenced using modern high-throughput DNA sequencers, and these sequences can be analysed with computer programs which quantify the recurrence of individual tags.
Analysis
The output of SAGE is a list of short sequence tags and the number of times it is observed. Using sequence databases a researcher can usually determine, with some confidence, from which original mRNA (and therefore which gene) the tag was extracted.
Statistical methods can be applied to tag and count lists from different samples in order to determine which genes are more highly expressed. For example, a normal tissue sample can be compared against a corresponding tumor to determine which genes tend to be more (or less) active.
History
In 1979 teams at Harvard and Caltech extended the basic idea of making DNA copies of mRNAs in vitro to amplifying a library of such in bacterial plasmids.[5] In 1982–1983, the idea of selecting random or semi-random clones from such a cDNA library for sequencing was explored by Greg Sutcliffe and coworkers.[6] and Putney et al. who sequenced 178 clones from a rabbit muscle cDNA library.[7] In 1991 Adams and co-workers coined the term expressed sequence tag (EST) and initiated more systematic sequencing of cDNAs as a project (starting with 600 brain cDNAs).[8] The identification of ESTs proceeded rapidly, millions of ESTs now available in public databases (e.g. GenBank).
In 1995, the idea of reducing the tag length from 100 to 800 bp down to tag length of 10 to 22 bp helped reduce the cost of mRNA surveys.[9] In this year, the original SAGE protocol was published by Victor Velculescu at the Oncology Center of Johns Hopkins University.[9] Although SAGE was originally conceived for use in cancer studies, it has been successfully used to describe the transcriptome of other diseases and in a wide variety of organisms.
Comparison to DNA microarrays
The general goal of the technique is similar to the DNA microarray. However, SAGE sampling is based on sequencing mRNA output, not on hybridization of mRNA output to probes, so transcription levels are measured more quantitatively than by microarray. In addition, the mRNA sequences do not need to be known a priori, so genes or gene variants which are not known can be discovered. Microarray experiments are much cheaper to perform, so large-scale studies do not typically use SAGE. Quantifying gene expressions is more exact in SAGE because it involves directly counting the number of transcripts whereas spot intensities in microarrays fall in non-discrete gradients and are prone to background noise.
Variant protocols
miRNA cloning
MicroRNAs, or miRNAs for short, are small (~22nt) segments of RNA which have been found to play a crucial role in gene regulation. One of the most commonly used methods for cloning and identifying miRNAs within a cell or tissue was developed in the Bartel Lab and published in a paper by Lau et al. (2001). Since then, several variant protocols have arisen, but most have the same basic format. The procedure is quite similar to SAGE: The small RNA are isolated, then linkers are added to each, and the RNA is converted to cDNA by RT-PCR. Following this, the linkers, containing internal restriction sites, are digested with the appropriate restriction enzyme and the sticky ends are ligated together into concatamers. Following concatenation, the fragments are ligated into plasmids and are used to transform bacteria to generate many copies of the plasmid containing the inserts. Those may then be sequenced to identify the miRNA present, as well as analysing expression levels of a given miRNA by counting the number of times it is present, similar to SAGE.
LongSAGE and RL-SAGE
LongSAGE was a more robust version of the original SAGE developed in 2002 which had a higher throughput, using 20 μg of mRNA to generate a cDNA library of thousands of tags.[10] Robust LongSage (RL-SAGE) Further improved on the LongSAGE protocol with the ability to generate a library with an insert size of 50 ng mRNA, much smaller than previous LongSAGE insert size of 2 μg mRNA[10] and using a lower number of ditag polymerase chain reactions (PCR) to obtain a complete cDNA library.[11]
SuperSAGE
SuperSAGE is a derivative of SAGE that uses the type III-endonuclease EcoP15I of phage P1, to cut 26 bp long sequence tags from each transcript's cDNA, expanding the tag-size by at least 6 bp as compared to the predecessor techniques SAGE and LongSAGE.[12] The longer tag-size allows for a more precise allocation of the tag to the corresponding transcript, because each additional base increases the precision of the annotation considerably.
Like in the original SAGE protocol, so-called ditags are formed, using blunt-ended tags. However, SuperSAGE avoids the bias observed during the less random LongSAGE 20 bp ditag-ligation.[13] By direct sequencing with high-throughput sequencing techniques (next-generation sequencing, i.e. pyrosequencing), hundred thousands or millions of tags can be analyzed simultaneously, producing very precise and quantitative gene expression profiles. Therefore, tag-based gene expression profiling also called "digital gene expression profiling" (DGE) can today provide most accurate transcription profiles that overcome the limitations of microarrays.[14][15]
3'end mRNA sequencing, massive analysis of cDNA ends
In the mid 2010s several techniques combined with Next Generation Sequencing were developed that employ the "tag" principle for "digital gene expression profiling" but without the use of the tagging enzyme. The "MACE" approach, (=Massive Analysis of cDNA Ends) generates tags somewhere in the last 1500 bps of a transcript. The technique does not depend on restriction enzymes anymore and thereby circumvents bias that is related to the absence or location of the restriction site within the cDNA. Instead, the cDNA is randomly fragmented and the 3'ends are sequenced from the 5' end of the cDNA molecule that carries the poly-A tail. The sequencing length of the tag can be freely chosen. Because of this, the tags can be assembled into contigs and the annotation of the tags can be drastically improved. Therefore, MACE is also use for the analyses of non-model organisms. In addition, the longer contigs can be screened for polymorphisms. As UTRs show a large number of polymorphisms between individuals, the MACE approach can be applied for allele determination, allele specific gene expression profiling and the search for molecular markers for breeding. In addition, the approach allows determining alternative polyadenylation of the transcripts. Because MACE does only require 3’ ends of transcripts, even partly degraded RNA can be analyzed with less degradation dependent bias. The MACE approach uses unique molecular identifiers to allow for identification of PCR bias. [16]
See also
References
- ^ Shafee, Thomas; Lowe, Rohan (2017). "Eukaryotic and prokaryotic gene structure". WikiJournal of Medicine. 4 (1). doi:10.15347/wjm/2017.002. ISSN 2002-4436.
- ^ Saha S, et al. (2002). "Using the transcriptome to annotate the genome". Nat Biotechnol. 20 (5): 508–12. doi:10.1038/nbt0502-508. PMID 11981567. S2CID 12709815.
- ^ Gowda M; Jantasuriyarat C; Dean RA; Wang GL. (2004). "Robust-LongSAGE (RL-SAGE): a substantially improved LongSAGE method for gene discovery and transcriptome analysis". Plant Physiol. 134 (3): 890–7. doi:10.1104/pp.103.034496. PMC 389912. PMID 15020752.
- ^ Matsumura H; Ito A; Saitoh H; Winter P; Kahl G; Reuter M; Krüger DH; Terauchi R. (2005). "SuperSAGE". Cell Microbiol. 7 (1): 11–8. doi:10.1111/j.1462-5822.2004.00478.x. PMID 15617519. S2CID 221579149.
- ^ Sim GK; Kafatos FC; Jones CW; Koehler MD; Efstratiadis A; Maniatis T (December 1979). "Use of a cDNA library for studies on evolution and developmental expression of the chorion multigene families". Cell. 18 (4): 1303–16. doi:10.1016/0092-8674(79)90241-1. PMID 519770.
- ^ Sutcliffe JG; Milner RJ; Bloom FE; Lerner RA (August 1982). "Common 82-nucleotide sequence unique to brain RNA". Proc Natl Acad Sci U S A. 79 (16): 4942–6. Bibcode:1982PNAS...79.4942S. doi:10.1073/pnas.79.16.4942. PMC 346801. PMID 6956902.
- ^ Putney SD; Herlihy WC; Schimmel P (1983). "A new troponin T and cDNA clones for 13 different muscle proteins, found by shotgun sequencing". Nature. 302 (5910): 718–21. Bibcode:1983Natur.302..718P. doi:10.1038/302718a0. PMID 6687628. S2CID 4364361.
- ^ Adams MD, Kelley JM, Gocayne JD, et al. (June 1991). "Complementary DNA sequencing: expressed sequence tags and human genome project". Science. 252 (5013): 1651–6. Bibcode:1991Sci...252.1651A. doi:10.1126/science.2047873. PMID 2047873. S2CID 13436211.
- ^ a b Velculescu VE; Zhang L; Vogelstein B; Kinzler KW. (1995). "Serial analysis of gene expression". Science. 270 (5235): 484–7. Bibcode:1995Sci...270..484V. doi:10.1126/science.270.5235.484. PMID 7570003. S2CID 16281846.
- ^ a b Saha, S., et al. (2002). "Using the transcriptome to annotate the genome." Nat Biotechnol 20(5): 508-512.
- ^ Gowda, M., et al. (2004). "Robust-LongSAGE (RL-SAGE): a substantially improved LongSAGE method for gene discovery and transcriptome analysis." Plant Physiol 134(3): 890-897.
- ^ Matsumura, H.; Reich, S.; Ito, A.; Saitoh, H.; Kamoun, S.; Winter, P.; Kahl, G.; Reuter, M.; Krüger, D.; Terauchi, R. (2003). "Gene expression analysis of plant host-pathogen interactions by SuperSAGE". Proceedings of the National Academy of Sciences. 100 (26): 15718–15723. Bibcode:2003PNAS..10015718M. doi:10.1073/pnas.2536670100. PMC 307634. PMID 14676315.
- ^ Gowda, Malali; Jantasuriyarat, Chatchawan; Dean, Ralph A.; Wang, Guo-Liang (2004-03-01). "Robust-LongSAGE (RL-SAGE): A Substantially Improved LongSAGE Method for Gene Discovery and Transcriptome Analysis". Plant Physiology. 134 (3): 890–897. doi:10.1104/pp.103.034496. ISSN 1532-2548. PMC 389912. PMID 15020752.
- ^ Shendure, J. (2008). "The beginning of the end for microarrays?". Nature Methods. 5 (7): 585–7. doi:10.1038/nmeth0708-585. PMID 18587314. S2CID 29682662.
- ^ Matsumura, H.; Bin Nasir, K. H.; Yoshida, K.; Ito, A.; Kahl, G. N.; Krüger, D. H.; Terauchi, R. (2006). "SuperSAGE array: the direct use of 26-base-pair transcript tags in oligonucleotide arrays". Nature Methods. 3 (6): 469–74. doi:10.1038/nmeth882. PMID 16721381. S2CID 19160070.
- ^ Zawada, Adam (January 2014). "Massive analysis of cDNA Ends (MACE) and miRNA expression profiling identifies proatherogenic pathways in chronic kidney disease". Epigenetics. 9 (1): 161–172. doi:10.4161/epi.26931. PMC 3928179. PMID 24184689.