RNF128
| RNF128 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | RNF128, GRAIL, ring finger protein 128, E3 ubiquitin protein ligase, ring finger protein 128 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 300439 MGI: 1914139 HomoloGene: 11337 GeneCards: RNF128 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
E3 ubiquitin-protein ligase RNF128 is an enzyme that in humans is encoded by the RNF128 gene.[5]
The protein encoded by this gene is a type I transmembrane protein that localizes to the endocytic pathway. This protein contains a RING zinc-finger motif and has been shown to possess E3 ubiquitin ligase activity. Expression of this gene in retrovirally transduced T cell hybridoma significantly inhibits activation-induced IL2 and IL4 cytokine production. Induced expression of this gene was observed in anergic CD4+ T cells, which suggested a role in the induction of anergic phenotype. Alternatively spliced transcript variants encoding distinct isoforms have been reported.[5] E3 ubiquitin-protein ligase RNF128 is highly expressed in the liver, adrenal glands, and intestines and also has notable expression in the kidneys, stomach, bladder, and thyroid. This protein lies in the endocytic pathway and contains a signal peptide, a RING zinc-finger motif, a protease associated domain, and a transmembrane domain.
Gene
RNF128 goes by other aliases including Gene Related to Anergy in Lymphocytes protein (GRAIL), E3 ubiquitin-protein ligase RNF128, FLJ23516, and RING finger protein 128.[6] The human RNF128 gene is located at Xq22.3 on the plus strand of the X chromosome and contains 8 exons and 7 introns.[5] This gene also has 234 orthologs in a span of organisms and is conserved in animals up to bony fish. Paralogs for this gene include RNF133, RNF150, RNF148, RNF149, RNF130, RNF13, RNF167, RNF215, and ZNRF4.
Transcript (mRNA)
Isoforms
RNF128 has two reported alternatively spliced transcript variants encoding isoforms. Isoform 1 contains 428 amino acids and isoform 2 contains 422 amino acids.[7] Isoform 1 has a longer transcript. Isoform 2 has an alternative 5' UTR and a different exon one compared to isoform 1. This results in a much shorter N-terminus in isoform 2. Isoform 1 is used more often when analyzing the protein. Isoform 1 includes a signal peptide, protease associated domain, transmembrane domain, and a RING zinc finger domain. Isoform 2 does not contain the same signal peptide or protease associated domain, but does include a similar transmembrane domain and RING zinc finger domain.
Protein
General properties
The RNF128 gene encodes and type I transmembrane protein. This protein functions as an E3 ubiquitin protein ligase that catalyzes Lys-43 and Lys-63 linked polyubiquitin chains and acts as an inhibitor of cytokine gene transcription when expressed in retrovirally transduced T cells.[8] This protein contains 428 amino acids and has two known isoforms.
Structure
RNF128 contains a N-terminal PA domain (residues 75–183) and a C-terminal RING finger domain (residues 277–318).[9] A crystallographic structure of the PA domain has been determined.[10]
Gene Level Regulation
Promoter
There are two total promoters, but the main promoter (GXP_14319) for RNF128 is 1076 nucleotides long. The transcription start site for RNF128 resides at the very end of the promoter sequence in the last 40 amino acids.[11]
Transcription Binding sites
There are many transcription factors that have a high affinity for binding RNF128's 5' UTR. Some important ones to mention are NFAT, a nuclear factor of activated T cells, EGRF, a Wilms tumor suppressor, and HNF6, a liver enriched cut-homeodomain transcription factor.[12]
Expression
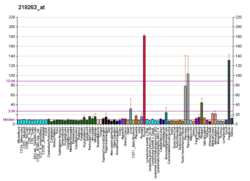
RNF128's expression in human tissues is very specific to the gut. There is very high expression in the liver and fetal liver especially. There is also high expression in the kidneys, adrenal glands, thyroid, small intestines and stomach.[13]
Transcript Level Regulation
There are over 10 stem loops in the 5' untranslated region on RNF128. There are also several areas of the 5' untranslated region that are highly conserved.[14]
Protein Level Regulation
The RNF128 protein contains a signal peptide. This peptide is cleaved at the RGA site 37 amino acids into the protein.[15] Myristoylation sites are predicted when the first 5 amino acids are removed from the sequence.[16] RNF128 also has three palmitoylation sites.[17] Myristoylation and palmitoylation adds a myristoyl and palmitoyl group to the protein which contrasts with N glycosylation because you are adding a hydrophobic tail. There are 6 predicted O glycosylation sites within this protein.[18] Out of the 6, only one of these O glycosylation sites are likely to be present as RNF128 is secreted. At O glycosylation sites, serines and threonines can be both phosphorylated and glycosylated and under different conditions, can be turned on or off. There are many phosphorylation sites for this protein, most of these being serines and a few threonine and tyrosine.[19] Phosphorylation is on the inside of the cell and can often turn certain signals on or off and can even lead to conformational changes in proteins. There are three significant sites for N glycosylation in this protein.[20] This could possibly protect the protein because of the large sugar complex and attract lectins that bind other proteins. Sugars from this N glycosylation also can change the shape of the protein which also helps it bind to other factors. The RNF128 protein has three different sumoylation sites.[21] These sites are similar to ubiquination in that they help target proteins for degradation under the right conditions. This helps the protein get rid of unwanted or needed regions. Research found that one third of the time this protein is found in the endoplasmic reticulum, one third in the plasma membrane, and the other third in the golgi.[22] This protein is considered to be localized in the endocytic pathway.

Homology & Evolution
Paralogs
Below is a table of RNF128 paralogs. Although there are many other paralogs than just these nine, these paralogs are the most closely related to RNF128.
| Paralog | E-value | Similarity % | Identity % | Relatedness |
| RNF133 | 4e-118 | 58 | 44 | Closely related |
| RNF150 | 9e-84 | 52 | 38 | Mod. related |
| RNF148 | 1e-99 | 49 | 37 | Mod. related |
| RNF149 | 2e-76 | 49 | 34 | Mod. related |
| RNF130 | 2e-72 | 46 | 33 | Mod. related |
| RNF13 | 4e-15 | 37 | 21 | Distantly related |
| RNF167 | 5e-14 | 35 | 21 | Distantly related |
| RNF215 | 1e-11 | 27 | 18 | Distantly related |
| ZNRF4 | 8e-10 | 33 | 19 | Distantly related |
Table 1: This table gives a list of nine RNF128 paralogs. Percent identity and percent similarity were found using EMBOSS Needle. The relatedness column gives insight into how closely related, moderately related, or distantly related that paralog is to RNF128.
Orthologs
RNF128 has 234 orthologs and is conserved in animals such as a dog, cow, mouse, rat, chicken, and a zebrafish. The most closely related orthologs reside in mammals with similarities between 75 and 100 percent. Moderately related orthologs resided in reptiles and birds with similarities between 67 and 75 percent. Then finally the most distantly related orthologs are amphibians and bony fish with similarity values around 60 percent. Many multiple sequence alignments were run using EMBOSS Global to look at the conservation of amino acids over time. The multiple sequence alignments compared distantly related and closely related homologs of RNF128. Many regions of RNF128 are conserved in the all species from mammals to bony fish, including the protease associated domain, transmembrane region, and the ring-H2 region. The signal peptide is not conserved in any of the more distant homologs, but is conserved in the strict orthologs. The ring-H2 region is the most highly conserved region in these alignments and is conserved in mammals, birds, amphibians, reptiles, and bony fish. Table 2 shows the phylogenetic tree of RNF128's orthologs.[23]

Evolution
RNF128 goes back approximately 433 m.y. The oldest life forms I found the gene to appear in are bony fish like zebrafish. This gene was not found in any invertebrates, fungus, bacteria, etc. The size of the gene family is 2. There are two isoforms produced by alternative splicing for RNF128 and I only found one splice isoform for the most distantly related organism. Figure 1 below shows the divergence of RNF128 over time and how slowly or quickly it diverged between organisms. When comparing my gene to cytochrome c and fibrinogen alpha, it can be determined that RNF128 diverges moderately slow. I used the slope of each line to find that a 1% change takes 27.7 million years for cytochrome c, 6.9 million years for RNF128, and 2.7 million years for fibrinogen alpha.

Interactions
RNF128 has been shown to interact with CD154[24] and OTUB1.[25] RNF128 interacts with many other different proteins including CD81, TP53, USP8, USP7, TBK1, and CD151.[26] The NSP7+NSP8 hexadecamer super complex is a SARS-Coronavirus RNA polymerase that interacts with RNF128. The NSP7+NSP8 super complex is heavily involved viral replication.[27]
Clinical significance
In multiple studies, RNF128 is associated with p53, a tumor suppressing gene. RNF128 is seen to act negatively on P53. Downregulation of RNF128 in certain studies leads to metastasis and a high mitotic rate in bladder and urothelial tissue.[28] Overexpression of RNF128 can inhibit p53-induced apoptosis by degradation of p53 and thus can be linked to a regulatory mechanism for the control of p53 under stressful circumstances.[29] RNF128 plays a role in CD4 and CD83 expression. It is able to down regulate the expression of CD83 on CD4 T cells.[30] RNF128 expression also limits IL2 and IL4 production by T lymphocytes.
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000133135 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000031438 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c "Entrez Gene: RNF128 ring finger protein 128".
- ^ "RNF128". Wikigenes.
- ^ "RNF128". NCBI protein.
- ^ "RNF128". Gene cards.
- ^ "Q8TEB7 (RN128_HUMAN)". UniProt.
- ^ "Protease-associated domain of the E3 ligase grail". RCSB Protein Databank.
- ^ Genomatix- https://www.genomatix.de/cgi-bin/dialign_prof/dialign.pl?s=11372be0a40d4bdcc5c71957820d9fb7;TASK=dialign_TF;SHOW=result_orthologs_Region_1_orthologs.seq.html
- ^ Genomatix- https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=449ecd52e5f13862195b729110712de2;PROM_ID=GXP_14319;GROUP=vertebrates;GROUP=others;ELDORADO_VERSION=E35R1911
- ^ Fagerberg L, Hallström BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397-406. doi:10.1074/mcp.M113.035600
- ^ http://unafold.rna.albany.edu/
- ^ SignalP- http://www.cbs.dtu.dk/cgi-bin/webface2.fcgi?jobid=5F0DD7E600002CE72D6D371A&wait=20
- ^ Myristoylator- https://web.expasy.org/cgi-bin/myristoylator/myristoylator.pl
- ^ CSS-Palm- http://csspalm.biocuckoo.org/showResult.php Archived 2018-07-20 at the Wayback Machine
- ^ YinOYang- http://www.cbs.dtu.dk/cgi-bin/webface2.fcgi?jobid=5F0DD7FA00002CE7D7515FD9&wait=20
- ^ NetPhos- http://www.cbs.dtu.dk/cgi-bin/webface2.fcgi?jobid=5F07487400006C9E3D0E94A1&wait=20
- ^ NetNGlyc- http://www.cbs.dtu.dk/cgi-bin/webface2.fcgi?jobid=5F0DD4DB0000662F2F470DB9&wait=20
- ^ GPS-SUMO- http://sumosp.biocuckoo.org/showResult.php Archived 2018-05-06 at the Wayback Machine
- ^ Psort-https://psort.hgc.jp/cgi-bin/runpsort.pl
- ^ Phylogeny Analysis- http://www.phylogeny.fr/phylogeny.cgi
- ^ Lineberry NB, Su LL, Lin JT, Coffey GP, Seroogy CM, Fathman CG (August 2008). "Cutting edge: The transmembrane E3 ligase GRAIL ubiquitinates the costimulatory molecule CD40 ligand during the induction of T cell anergy". Journal of Immunology. 181 (3): 1622–6. doi:10.4049/jimmunol.181.3.1622. PMC 2853377. PMID 18641297.
- ^ Soares L, Seroogy C, Skrenta H, Anandasabapathy N, Lovelace P, Chung CD, et al. (January 2004). "Two isoforms of otubain 1 regulate T cell anergy via GRAIL". Nature Immunology. 5 (1): 45–54. doi:10.1038/ni1017. PMID 14661020. S2CID 27005972.
- ^ Mentha- http://mentha.uniroma2.it/result.php#RNF128
- ^ NCBI- https://www.ncbi.nlm.nih.gov/Structure/cdd/PF08716
- ^ Lee YY, Wang CT, Huang SK, Wu WJ, Huang CN, Li CC, et al. (2016). "Downregulation of RNF128 Predicts Progression and Poor Prognosis in Patients with Urothelial Carcinoma of the Upper Tract and Urinary Bladder". Journal of Cancer. 7 (15): 2187–2196. doi:10.7150/jca.16798. PMC 5166527. PMID 27994654.
- ^ Chen YC, Chan JY, Chiu YL, Liu ST, Lozano G, Wang SL, et al. (May 2013). "Grail as a molecular determinant for the functions of the tumor suppressor p53 in tumorigenesis". Cell Death and Differentiation. 20 (5): 732–43. doi:10.1038/cdd.2013.1. PMC 3619241. PMID 23370271.
- ^ Su LL, Iwai H, Lin JT, Fathman CG (July 2009). "The transmembrane E3 ligase GRAIL ubiquitinates and degrades CD83 on CD4 T cells". Journal of Immunology. 183 (1): 438–44. doi:10.4049/jimmunol.0900204. PMC 4300110. PMID 19542455.
Further reading
- Kostianovsky AM, Maier LM, Baecher-Allan C, Anderson AC, Anderson DE (May 2007). "Up-regulation of gene related to anergy in lymphocytes is associated with Notch-mediated human T cell suppression". Journal of Immunology. 178 (10): 6158–63. doi:10.4049/jimmunol.178.10.6158. PMID 17475842.
- MacKenzie DA, Schartner J, Lin J, Timmel A, Jennens-Clough M, Fathman CG, Seroogy CM (March 2007). "GRAIL is up-regulated in CD4+ CD25+ T regulatory cells and is sufficient for conversion of T cells to a regulatory phenotype". The Journal of Biological Chemistry. 282 (13): 9696–702. doi:10.1074/jbc.M604192200. PMID 17259178.
- Anandasabapathy N, Ford GS, Bloom D, Holness C, Paragas V, Seroogy C, et al. (April 2003). "GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells". Immunity. 18 (4): 535–47. doi:10.1016/S1074-7613(03)00084-0. PMID 12705856.
External links
- Webarchive template wayback links
- Articles with short description
- Short description matches Wikidata
- Articles needing cleanup from August 2022
- Articles with bare URLs for citations from August 2022
- All articles with bare URLs for citations
- Articles covered by WikiProject Wikify from August 2022
- All articles covered by WikiProject Wikify
- Genes on human chromosome X
- RING finger proteins