KLK6
| KLK6 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | KLK6, Bssp, Klk7, PRSS18, PRSS9, SP59, hK6, kallikrein related peptidase 6 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 602652 MGI: 1343166 HomoloGene: 68279 GeneCards: KLK6 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Kallikrein-6 is a protein that in humans is encoded by the KLK6 gene.[5][6][7][8] Kallikrein-6 is also referred to as neurosin, protease M, hK6, or zyme. It is a 223 amino acid sequence, derived from its 244 original form, which contains a 16 residue presignal and 5 residue activation peptide.[9]
Function
Kallikreins are a subgroup of serine proteases having diverse physiological functions. Growing evidence suggests that many kallikreins are implicated in carcinogenesis and some have potential as novel cancer and other disease biomarkers. This gene is one of the fifteen kallikrein subfamily members located in a cluster on chromosome 19. The encoded enzyme is regulated by steroid hormones. In tissue culture, the enzyme has been found to generate amyloidogenic fragments from the amyloid precursor protein, suggesting a potential for involvement in Alzheimer's disease. Multiple alternatively spliced transcript variants that encode different isoforms have been identified for this gene.[8]
Structure
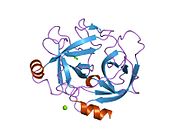
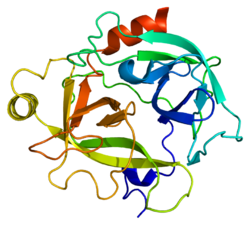
The secondary structure consists of 13 beta-pleated sheets, 2 alpha-helices, 2 310-helices, and 8 loop regions. In terms of amino acid sequences, hK6 is most similar to myelencephalon-specific protease (MSP), which comes from the rat kvllikrein gene family. MSP and hK6 both target the peptide bond where arginine follows and they both automatically cleave themselves at their Arg positions.[10]
However, structurally, hK6 most resembles trypsin found in cows/oxen. Surrounding the active site, there are short loop regions that point away from the binding site. In the binding site, residues 189-195, 214-220, and 224-228 are found in addition to the Asp, His, and Ser residues.[11]
Disease Pathology
Alpha-synuclein build-up is commonly found in Dementia with Lewy bodies, Parkinson's disease, and multiple system atrophy patients, serving as a biomarker for infection. Thus, degradation of this protein is necessary to prevent infection. In mice brain samples, protease inhibitors were used to identify the protein responsible for alpha-synuclein degradation. Various serine protease inhibitors (aprotinin, phenylmethyl sulfonyl fluoride, leupeptin, and 4-(2-aminoethyl)-benzenesulfonyl fluoride). significantly affected the degradation pathway, which justifies the necessity for a serine protease to degrade alpha-synuclein.
Kallikrein inhibitor was introduced to the mice samples, and it successfully inhibited kallikrein function. In vitro studies utilizing purified kallikrein were also performed on alpha-synuclein, and it was effective in degrading alpha-synuclein. Both the inhibition and successful in vitro enzymatic activity demonstrates kallikrein as the degradation enzyme.[12]
While hK6 has contributed to disease prevention, it also has the potential to contribute to the spread of malignant tumor cells. As a degradation enzyme, it has the capability of degrading extracellular matrix proteins on both normal and malignant cells, which would enhance their abilities to migrate and to send signals. For example, fibronectin interacts with integral molecules as malignant cells try to migrate; by degrading it, malignant cells are able to migrate, attach, and send a signal to other malignant cells.[13]
Neurosin Mechanism

Asp-102, His-57, and Ser-195 form a catalytic triad to specifically hydrolyze a peptide bond where Arg is towards the N terminus and a general amino acid is towards the C terminus.[14] It is believed to follow a similar pathway to other serine-type proteases.[15]
- Histidine deprotonates serine
- Serine substitutes in at the amide bond
- The protonated histidine makes the amine a better leaving group and the oxyanion collapses to form the ester.
- Water enters the triad and cleaves the ester bond, releasing serine.
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000167755 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000050063 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Yamashiro K, Tsuruoka N, Kodama S, Tsujimoto M, Yamamura Y, Tanaka T, et al. (January 1997). "Molecular cloning of a novel trypsin-like serine protease (neurosin) preferentially expressed in brain". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1350 (1): 11–14. doi:10.1016/s0167-4781(96)00187-x. PMID 9003450.
- ^ Lundwall A, Band V, Blaber M, Clements JA, Courty Y, Diamandis EP, et al. (June 2006). "A comprehensive nomenclature for serine proteases with homology to tissue kallikreins" (PDF). Biological Chemistry. 387 (6): 637–641. doi:10.1515/BC.2006.082. PMID 16800724. S2CID 436200.
- ^ "Proceedings of the 1st International Symposium on Kallikreins, Lausanne, Switzerland, September 1-3 , 2005". Biological Chemistry. 387 (6): 635–824. June 2006. doi:10.1515/BC.2006.081. PMID 16800723. S2CID 83910246.
- ^ a b "Entrez Gene: KLK6 kallikrein-related peptidase 6".
- ^ Gomis-Rüth FX, Bayés A, Sotiropoulou G, Pampalakis G, Tsetsenis T, Villegas V, et al. (July 2002). "The structure of human prokallikrein 6 reveals a novel activation mechanism for the kallikrein family". The Journal of Biological Chemistry. 277 (30): 27273–27281. doi:10.1074/jbc.M201534200. PMID 12016211.
- ^ Bernett MJ, Blaber SI, Scarisbrick IA, Dhanarajan P, Thompson SM, Blaber M (July 2002). "Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system". The Journal of Biological Chemistry. 277 (27): 24562–24570. doi:10.1074/jbc.M202392200. PMID 11983703.
- ^ Bernett MJ, Blaber SI, Scarisbrick IA, Dhanarajan P, Thompson SM, Blaber M (July 2002). "Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system". The Journal of Biological Chemistry. 277 (27): 24562–24570. doi:10.1074/jbc.M202392200. PMID 11983703.
- ^ Iwata A, Maruyama M, Akagi T, Hashikawa T, Kanazawa I, Tsuji S, Nukina N (October 2003). "Alpha-synuclein degradation by serine protease neurosin: implication for pathogenesis of synucleinopathies". Human Molecular Genetics. 12 (20): 2625–2635. doi:10.1093/hmg/ddg283. PMID 12928483.
- ^ Ghosh MC, Grass L, Soosaipillai A, Sotiropoulou G, Diamandis EP (2004). "Human kallikrein 6 degrades extracellular matrix proteins and may enhance the metastatic potential of tumour cells". Tumour Biology. 25 (4): 193–199. doi:10.1159/000081102. PMID 15557757. S2CID 14391147.
- ^ Blaber SI, Yoon H, Scarisbrick IA, Juliano MA, Blaber M (May 2007). "The autolytic regulation of human kallikrein-related peptidase 6". Biochemistry. 46 (17): 5209–5217. doi:10.1021/bi6025006. PMC 2517904. PMID 17417874.
- ^ Hedstrom L (December 2002). "Serine protease mechanism and specificity". Chemical Reviews. 102 (12): 4501–4524. doi:10.1021/cr000033x. PMID 12475199.
Further reading
- Yousef GM, Kishi T, Diamandis EP (March 2003). "Role of kallikrein enzymes in the central nervous system". Clinica Chimica Acta; International Journal of Clinical Chemistry. 329 (1–2): 1–8. doi:10.1016/S0009-8981(03)00004-4. PMID 12589961.
- Menendez-Gonzalez M, Castro-Santos P, Suarez A, Calatayud MT, Perez-Pinera P, Martinez M, et al. (May 2008). "Value of measuring plasmatic levels of neurosin in the diagnosis of Alzheimer's disease". Journal of Alzheimer's Disease. 14 (1): 59–67. doi:10.3233/JAD-2008-14106. PMID 18525128.
- Menendez-Gonzalez M, Castro-Santos P, Calatayud MT, Perez-Piñera P, Ribacoba R, Martinez-Rivera M, et al. (July 2008). "Plasmatic level of neurosin predicts outcome of mild cognitive impairment". International Archives of Medicine. 1 (1): 11. doi:10.1186/1755-7682-1-11. PMC 2475518. PMID 18620574.
- Anisowicz A, Sotiropoulou G, Stenman G, Mok SC, Sager R (September 1996). "A novel protease homolog differentially expressed in breast and ovarian cancer". Molecular Medicine. 2 (5): 624–636. doi:10.1007/BF03401646. PMC 2230195. PMID 8898378.
- Little SP, Dixon EP, Norris F, Buckley W, Becker GW, Johnson M, et al. (October 1997). "Zyme, a novel and potentially amyloidogenic enzyme cDNA isolated from Alzheimer's disease brain". The Journal of Biological Chemistry. 272 (40): 25135–25142. doi:10.1074/jbc.272.40.25135. PMID 9312124.
- Yousef GM, Luo LY, Scherer SW, Sotiropoulou G, Diamandis EP (December 1999). "Molecular characterization of zyme/protease M/neurosin (PRSS9), a hormonally regulated kallikrein-like serine protease". Genomics. 62 (2): 251–259. doi:10.1006/geno.1999.6012. PMID 10610719.
- Gan L, Lee I, Smith R, Argonza-Barrett R, Lei H, McCuaig J, et al. (October 2000). "Sequencing and expression analysis of the serine protease gene cluster located in chromosome 19q13 region". Gene. 257 (1): 119–130. doi:10.1016/S0378-1119(00)00382-6. PMID 11054574.
- Petraki CD, Karavana VN, Skoufogiannis PT, Little SP, Howarth DJ, Yousef GM, Diamandis EP (November 2001). "The spectrum of human kallikrein 6 (zyme/protease M/neurosin) expression in human tissues as assessed by immunohistochemistry". The Journal of Histochemistry and Cytochemistry. 49 (11): 1431–1441. doi:10.1177/002215540104901111. PMID 11668196.
- Bernett MJ, Blaber SI, Scarisbrick IA, Dhanarajan P, Thompson SM, Blaber M (July 2002). "Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system". The Journal of Biological Chemistry. 277 (27): 24562–24570. doi:10.1074/jbc.M202392200. PMID 11983703.
- Gomis-Rüth FX, Bayés A, Sotiropoulou G, Pampalakis G, Tsetsenis T, Villegas V, et al. (July 2002). "The structure of human prokallikrein 6 reveals a novel activation mechanism for the kallikrein family". The Journal of Biological Chemistry. 277 (30): 27273–27281. doi:10.1074/jbc.M201534200. PMID 12016211.
- Scarisbrick IA, Blaber SI, Lucchinetti CF, Genain CP, Blaber M, Rodriguez M (June 2002). "Activity of a newly identified serine protease in CNS demyelination". Brain. 125 (Pt 6): 1283–1296. doi:10.1093/brain/awf142. PMID 12023317.
- Zarghooni M, Soosaipillai A, Grass L, Scorilas A, Mirazimi N, Diamandis EP (May 2002). "Decreased concentration of human kallikrein 6 in brain extracts of Alzheimer's disease patients". Clinical Biochemistry. 35 (3): 225–231. doi:10.1016/S0009-9120(02)00292-8. PMID 12074831.
- Hoffman BR, Katsaros D, Scorilas A, Diamandis P, Fracchioli S, Rigault de la Longrais IA, et al. (September 2002). "Immunofluorometric quantitation and histochemical localisation of kallikrein 6 protein in ovarian cancer tissue: a new independent unfavourable prognostic biomarker". British Journal of Cancer. 87 (7): 763–771. doi:10.1038/sj.bjc.6600533. PMC 2364256. PMID 12232761.
- Mitsui S, Okui A, Uemura H, Mizuno T, Yamada T, Yamamura Y, Yamaguchi N (November 2002). "Decreased cerebrospinal fluid levels of neurosin (KLK6), an aging-related protease, as a possible new risk factor for Alzheimer's disease". Annals of the New York Academy of Sciences. 977 (1): 216–223. Bibcode:2002NYASA.977..216M. doi:10.1111/j.1749-6632.2002.tb04818.x. PMID 12480753. S2CID 40568196.
- Hutchinson S, Luo LY, Yousef GM, Soosaipillai A, Diamandis EP (May 2003). "Purification of human kallikrein 6 from biological fluids and identification of its complex with alpha(1)-antichymotrypsin". Clinical Chemistry. 49 (5): 746–751. doi:10.1373/49.5.746. PMID 12709365.
- Magklara A, Mellati AA, Wasney GA, Little SP, Sotiropoulou G, Becker GW, Diamandis EP (August 2003). "Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and regulation by inhibitors". Biochemical and Biophysical Research Communications. 307 (4): 948–955. doi:10.1016/S0006-291X(03)01271-3. PMID 12878203.
- Iwata A, Maruyama M, Akagi T, Hashikawa T, Kanazawa I, Tsuji S, Nukina N (October 2003). "Alpha-synuclein degradation by serine protease neurosin: implication for pathogenesis of synucleinopathies". Human Molecular Genetics. 12 (20): 2625–2635. doi:10.1093/hmg/ddg283. PMID 12928483.
- Sauter ER, Lininger J, Magklara A, Hewett JE, Diamandis EP (February 2004). "Association of kallikrein expression in nipple aspirate fluid with breast cancer risk". International Journal of Cancer. 108 (4): 588–591. doi:10.1002/ijc.11607. PMID 14696124. S2CID 29116787.
- Pampalakis G, Kurlender L, Diamandis EP, Sotiropoulou G (July 2004). "Cloning and characterization of novel isoforms of the human kallikrein 6 gene". Biochemical and Biophysical Research Communications. 320 (1): 54–61. doi:10.1016/j.bbrc.2004.04.205. PMID 15207701.