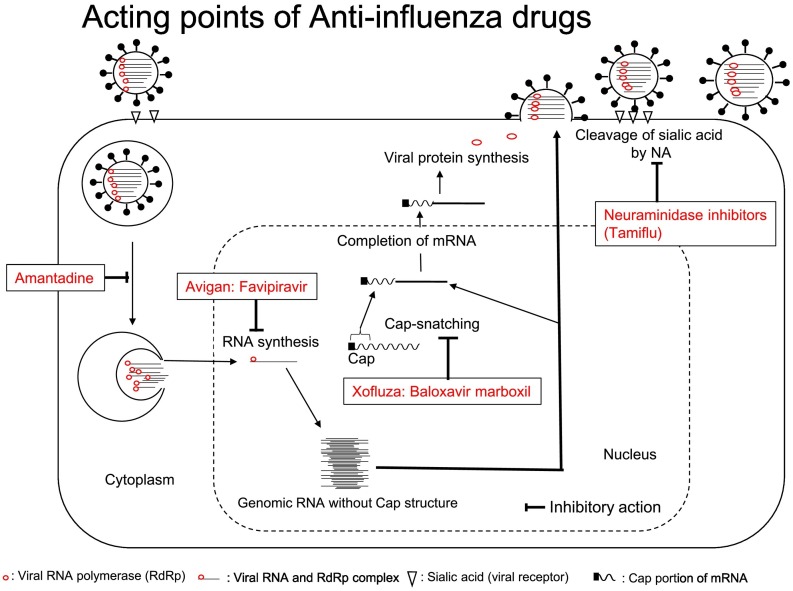
File:Gr4 lrgnih.jpg
Jump to navigation
Jump to search
Gr4_lrgnih.jpg (793 × 591 pixels, file size: 89 KB, MIME type: image/jpeg)
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 16:46, 20 May 2023 |  | 793 × 591 (89 KB) | Ozzie10aaaa | Uploaded a work by Kimiyasu Shirakia, Tohru Daikokub from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7102570/ with UploadWizard |
File usage
There are no pages that use this file.