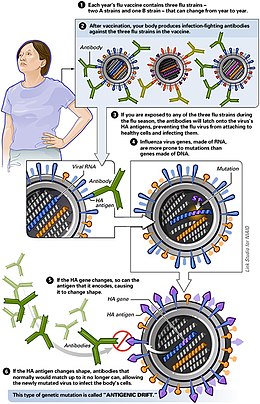
Antigenic drift

Antigenic drift is a kind of genetic variation in viruses, arising from the accumulation of mutations in the virus genes that code for virus-surface proteins that host antibodies recognize. This results in a new strain of virus particles that is not effectively inhibited by the antibodies that prevented infection by previous strains. This makes it easier for the changed virus to spread throughout a partially immune population. Antigenic drift occurs in both influenza A and influenza B viruses.[1][2][3][4]
Confusion can arise with two very similar terms, antigenic shift and genetic drift. Antigenic shift is a closely related process; it refers to more dramatic changes in the virus's surface proteins. Genetic drift is very different and much more broadly applicable; it refers to the gradual accumulation in any DNA sequence of random mutational changes that do not interfere with the DNA's function and thus that are not seen by natural selection.[5][6]
The immune system recognizes viruses when antigens on the surfaces of virus particles bind to immune receptors that are specific for these antigens. These receptors can be antibodies in the bloodstream or similar proteins on the surfaces of immune-system cells. After an infection or after vaccination, the body produces many more of these virus-specific immune receptors, which prevent re-infection by this particular strain of the virus; this is called acquired immunity. However, viral genomes are constantly mutating, producing new forms of these antigens. If one of these new forms of an antigen is sufficiently different from the old antigen, it will no longer bind to the antibodies or immune-cell receptors, allowing the mutant virus to infect people who were immune to the original strain of the virus because of prior infection or vaccination.[7][8][4]
In 1940s, Maurice Hilleman discovered antigenic drift, which is the most common way that influenza viruses change.[9][10][11][12] A second type of change is antigenic shift, also discovered by Hilleman,[9][10] where the virus acquires a completely new version of one of its surface-protein genes from a distantly related influenza virus. The rate of antigenic drift is dependent on two characteristics: the duration of the epidemic, and the strength of host immunity. A longer epidemic allows for selection pressure to continue over an extended period of time and stronger host immune responses increase selection pressure for development of novel antigens.[13]
In influenza viruses

In the influenza virus, the two relevant antigens are the surface proteins, hemagglutinin and neuraminidase.[14]
The hemagglutinin is responsible for binding and entry into host epithelial cells while the neuraminidase is involved in the process of new virions budding out of host cells.[15]
Sites recognized on the hemagglutinin and neuraminidase proteins by host immune systems are under constant selective pressure. Antigenic drift allows for evasion of these host immune systems by small mutations in the hemagglutinin and neuraminidase genes that make the protein unrecognizable to pre-existing host immunity.[16]
Antigenic drift is this continuous process of genetic and antigenic change among flu strains.[17]
In human populations, immune (vaccinated) individuals exert selective pressure for single point mutations in the hemagglutinin gene that increase receptor binding avidity, while naive individuals exert selective pressure for single point mutations that decrease receptor binding avidity.[16]
These dynamic selection pressures facilitate the observed rapid evolution in the hemagglutinin gene. Specifically, 18 specific codons in the HA1 domain of the hemagglutinin gene have been identified as undergoing positive selection to change their encoded amino acid.[18]
To meet the challenge of antigenic drift, vaccines that confer broad protection against heterovariant strains are needed against seasonal, epidemic and pandemic influenza.[19] As in all RNA viruses, mutations in influenza occur frequently because the virus' RNA polymerase has no proofreading mechanism, resulting in an error rate between 1×10−3 and 8×10−3 substitutions per site per year during viral genome replication.[17]
Mutations in the surface proteins allow the virus to elude some host immunity, and the numbers and locations of these mutations that confer the greatest amount of immune escape has been an important topic of study for over a decade.[20][21][22]
Antigenic drift has been responsible for heavier-than-normal flu seasons in the past, like the outbreak of influenza H3N2 variant A/Fujian/411/2002 in the 2003–2004 flu season. All influenza viruses experience some form of antigenic drift.[23][19]
Antigenic drift should not be confused with antigenic shift, which refers to reassortment of the virus' gene segments. As well, it is different from random genetic drift, which is an important mechanism in population genetics.[5][6][24]
-
Differences between Antigenic shift and Antigenic drift
-
Antigenic drift is the process by which flu virus genetically mutates, producing new virus strains that may not be recognozed by body's immune system.
See also
References
- ↑ Shors, Teri (2009). Understanding Viruses. Jones & Bartlett Learning. p. 327. ISBN 978-0-7637-2932-5. Archived from the original on 20 September 2022. Retrieved 15 September 2022.
- ↑ Treanor, John J. (1 January 2015). "167 - Influenza (Including Avian Influenza and Swine Influenza)". Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases (Eighth Edition). W.B. Saunders. pp. 2000–2024.e6. ISBN 978-1-4557-4801-3. Archived from the original on 27 April 2022. Retrieved 18 September 2022.
- ↑ Kurstak, Edouard; Marusyk, R. G.; Murphy, F. A.; Regenmortel, M. H. V. Van (11 November 2013). Virus Variability, Epidemiology and Control. Springer Science & Business Media. p. 4. ISBN 978-1-4757-9271-3.
- ↑ 4.0 4.1 Couch, Robert B. (1996). "Orthomyxoviruses". Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 978-0-9631172-1-2. Archived from the original on 2021-10-23. Retrieved 22 September 2022.
- ↑ 5.0 5.1 "How Flu Viruses Can Change". Centers for Disease Control and Prevention. 21 September 2021. Archived from the original on 18 March 2021. Retrieved 16 September 2022.
- ↑ 6.0 6.1 Masel, Joanna (25 October 2011). "Genetic drift". Current biology: CB. 21 (20): R837–838. doi:10.1016/j.cub.2011.08.007. ISSN 1879-0445. Archived from the original on 16 June 2022. Retrieved 18 September 2022.
- ↑ Mueller, Scott N.; Rouse, Barry T. (2008). "Immune responses to viruses". Clinical Immunology: 421–431. doi:10.1016/B978-0-323-04404-2.10027-2. Archived from the original on 2 August 2022. Retrieved 15 September 2022.
- ↑ Klimpel, Gary R. (1996). "Immune Defenses". Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 978-0-9631172-1-2. Archived from the original on 2022-08-14. Retrieved 17 September 2022.
- ↑ 9.0 9.1 Oransky, Ivan (2005-05-14). "Maurice R Hilleman". The Lancet. 365 (9472): 1682. doi:10.1016/S0140-6736(05)66536-1. ISSN 0140-6736. PMID 15912596. S2CID 46630955.
- ↑ 10.0 10.1 Kurth, Reinhard (April 2005). "Maurice R. Hilleman (1919–2005)". Nature. 434 (7037): 1083. doi:10.1038/4341083a. ISSN 1476-4687. PMID 15858560.
- ↑ D. J. D. Earn; J. Dushoff; S. A. Levin (2002). "Ecology and Evolution of the Flu". Trends in Ecology and Evolution. 17 (7): 334–340. doi:10.1016/S0169-5347(02)02502-8.
- ↑ A. W. Hampson (2002). "Influenza virus antigens and antigenic drift". In C. W. Potter (ed.). Influenza. Elsevier Science B. V. pp. 49–86. ISBN 978-0-444-82461-5.
- ↑ Boni, T; S. Cobey; P. Beerli; M. Pascual (2006). "Epidemic dynamics and antigenic evolution in a single season of influenza A". Proceedings of the Royal Society B. 273 (1592): 1307–1316. doi:10.1098/rspb.2006.3466. PMC 1560306. PMID 16777717.
- ↑ Bouvier NM, Palese P (Sep 2008). "The biology of influenza viruses". Vaccine. 26 (Suppl 4): D49–53. doi:10.1016/j.vaccine.2008.07.039. PMC 3074182. PMID 19230160.
- ↑ Nelson, M. I.; Holmes, E. C. (March 2007). "The evolution of pandemic influenza". Nature Reviews Genetics. 8 (3): 196–205. doi:10.1038/nrg2053. PMID 17262054. S2CID 221107.
- ↑ 16.0 16.1 Hensley, S. E.; Das, S. R.; Bailey, A. L.; Schmidt, L. M.; Hickman, H. D.; Jayaraman, A.; Viswanathan, K.; Raman, R.; Sasisekharan, R.; Bennink, J. R.; Yewdell, J. W. (30 October 2009). "Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift". Science. 326 (5953): 734–736. Bibcode:2009Sci...326..734H. doi:10.1126/science.1178258. PMC 2784927. PMID 19900932.
- ↑ 17.0 17.1 Taubenberger, Jeffery K.; Kash, John C. (17 June 2010). "Influenza virus evolution, host adaptation and pandemic formation". Cell Host & Microbe. 7 (6): 440–451. doi:10.1016/j.chom.2010.05.009. PMC 2892379. PMID 20542248.
- ↑ Bush, R. M.; K. Subbarao; N. J. Cox; W. M. Fitch (3 December 1999). "Predicting the evolution of human influenza A". Science. 286 (5446): 1921–1925. doi:10.1126/science.286.5446.1921. PMID 10583948. S2CID 2836600.
- ↑ 19.0 19.1 Carrat, F.; Flahault, A. (28 September 2007). "Influenza vaccine: the challenge of antigenic drift". Vaccine. 25 (39–40): 6852–6862. doi:10.1016/j.vaccine.2007.07.027. ISSN 0264-410X. Archived from the original on 8 February 2022. Retrieved 19 September 2022.
- ↑ R. M. Bush; W. M. Fitch; C. A. Bender; N. J. Cox (1999). "Positive selection on the H3 hemagglutinin gene of human influenza virus". Molecular Biology and Evolution. 16 (11): 1457–1465. doi:10.1093/oxfordjournals.molbev.a026057. PMID 10555276.
- ↑ W. M. Fitch; R. M. Bush; C. A. Bender; N. J. Cox (1997). "Long term trends in the evolution of H(3) HA1 human influenza type A". Proceedings of the National Academy of Sciences of the United States of America. 94 (15): 7712–7718. Bibcode:1997PNAS...94.7712F. doi:10.1073/pnas.94.15.7712. PMC 33681. PMID 9223253.
- ↑ D. J. Smith, A. S. Lapedes, J. C. de Jong, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. M. E. Osterhaus, R. A. M. Fouchier (2004). "Mapping the antigenic and genetic evolution of influenza virus" (PDF). Science. 305 (5682): 371–376. Bibcode:2004Sci...305..371S. doi:10.1126/science.1097211. PMID 15218094. S2CID 1258353. Archived from the original (PDF) on 2019-03-07.
{{cite journal}}: CS1 maint: uses authors parameter (link) - ↑ "Update: Influenza Activity --- United States and Worldwide, 2003--04 Season, and Composition of the 2004--05 Influenza Vaccine". web.archive.org. 4 September 2021. Archived from the original on 4 September 2021. Retrieved 17 September 2022.
{{cite web}}: CS1 maint: bot: original URL status unknown (link) - ↑ Chen, Yuxin; Tong, Ding; Wu, Chung-I. (1 August 2017). "A New Formulation of Random Genetic Drift and Its Application to the Evolution of Cell Populations". Molecular Biology and Evolution. 34 (8): 2057–2064. doi:10.1093/molbev/msx161. ISSN 1537-1719. Archived from the original on 21 June 2022. Retrieved 20 September 2022.
Further reading
- Boni MF (July 2008). "Vaccination and antigenic drift in influenza". Vaccine. 26 Suppl 3: C8–14. doi:10.1016/j.vaccine.2008.04.011. PMC 2603026. PMID 18773534.
- Gog JR (July 2008). "The impact of evolutionary constraints on influenza dynamics". Vaccine. 26 Suppl 3: C15–24. doi:10.1016/j.vaccine.2008.04.008. PMID 18773528.
External links
- An illustration of antigenic drift
- A technical definition Archived 2014-05-12 at the Wayback Machine